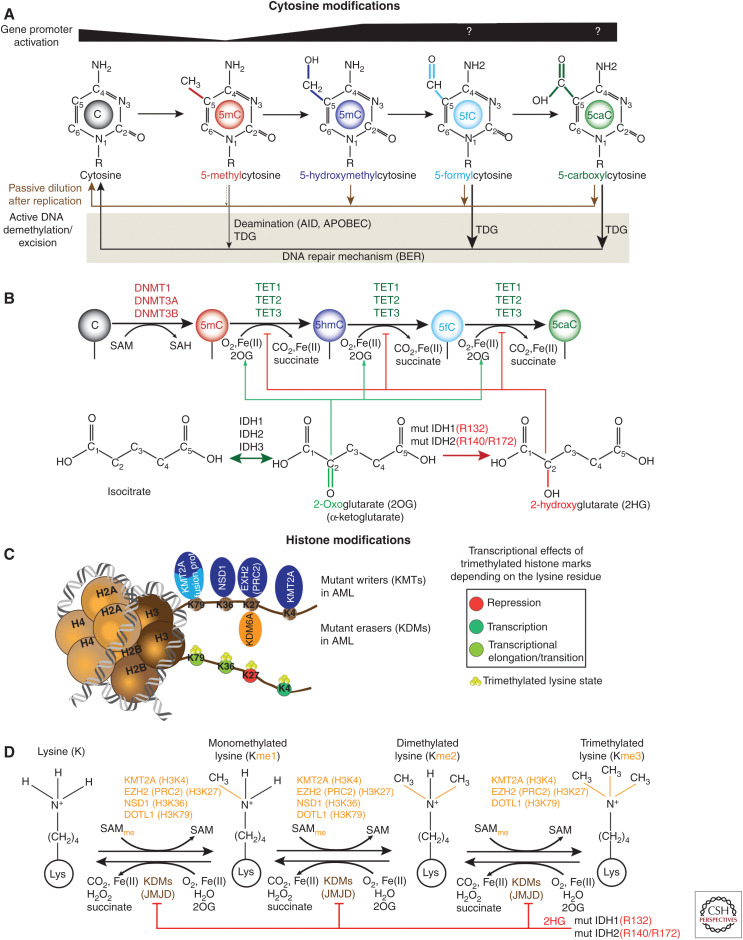
Figure 2.
Epigenetic features and perturbations in acute myeloid leukemia (AML). (A) Diagram showing the structure of methylated cytosine (5mC) and oxidized mCs (5hmC, 5fC, 5caC) as well as their transcriptional association at promoters. Active DNA demethylation is indicated by black arrows below modified cytosines. 5fC and 5caC can be excised directly by the thymine DNA glycosylase (TDG), resulting in an abasic site that is eventually replaced with unmethylated cytosine by the base excision repair (BER) machinery. Activation-induced cytidine deaminase (AID) or APOBEC can deaminated 5mC to thymine that is subsequently removed by TDG and repaired by BER. On the other hand, passive demethylation (brown arrows) is an alternative process that can occur through dilution following cell replication. DNMT1 can directly replicate the 5mC pattern onto the newly synthesized daughter strand unlike the pattern of oxidized mCs. (For review, see Rasmussen and Helin 2016.) (B) Enzymatic processes involved in cytosine methylation and oxidation of 5mC. DNMTs catalyze the addition of a methyl group to cytosine using S-adenosyl methionine (SAM) as methyl donor. Ten-eleven translocation (TET) proteins oxidize methylated cytosines to 5hmC, 5fC, and 5caC in an iterative manner using 2OG (αKG) as co-substrate. Mutant IDH1/2 generates 2HG that inhibits competitively the enzymatic activity of TET proteins. (C) Nucleosome model shows lysine substrate residues on histone H3 for writers (KMTs) and erasers (KDMs) that are found mutated in AML. Chromosomal translocations involving KMT2A (KMT2A-re) often result in fusion proteins that associate with the H3K79 histone methyltransferase DOT1L. (D) Enzymatic processes of lysine (de)methylation for selected KMTs. Besides TET proteins, 2HG can also impair the enzymatic activity Jumonji domain (JMJD)-containing KDMs.