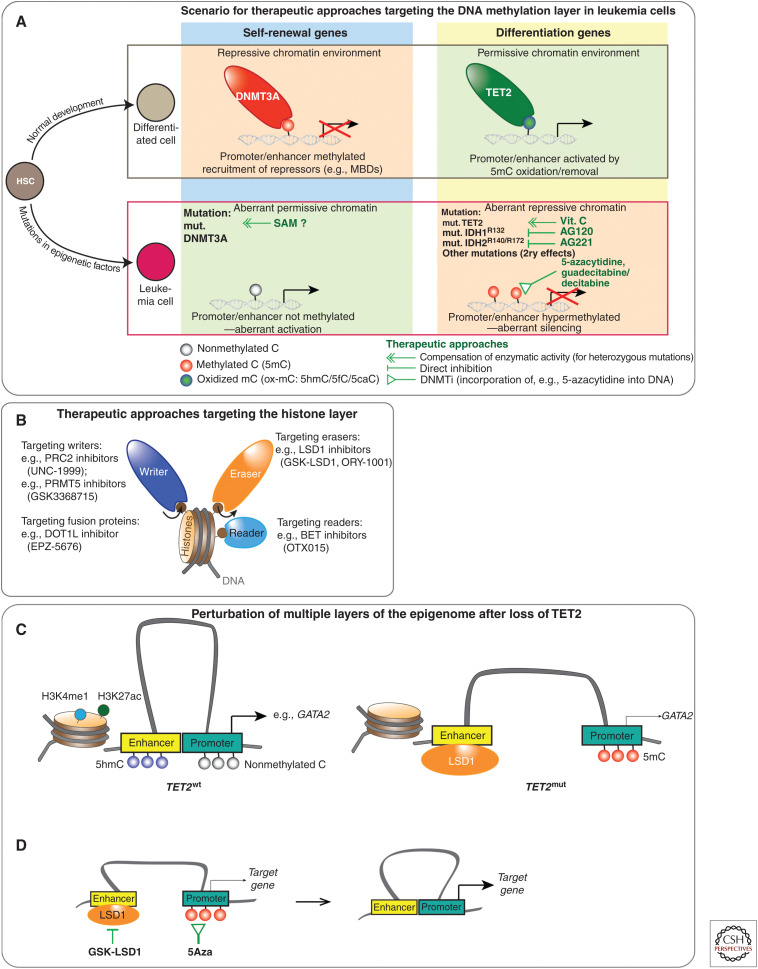
Figure 3.
Epigenetic therapy and combination therapy targeting cooperating layers of the epigenome in acute myeloid leukemia (AML). (A) Scenario illustrating the effect of distinct epigenetic mutations (indicated in bold letters) in leukemia cells versus normal differentiated cells derived from hematopoietic stem cells (HSCs). Epigenetic therapy and strategies targeting aberrant DNA methylation are shown by green lines. Loss of DNA methylation at self-renewal genes in DNMT3Amut AML may be potentially reversible by increasing intracellular levels of SAM (methyl donor). Loss of TET2 function can be compensated with vitamin C treatment. Specific inhibitors (AG120, AG221) against mutant IDH1/2 can block production of the TET inhibitor 2HG. Direct DNA hypermethylation (mutant TET2 or IDH1/2) or indirect hypermethylation due to secondary effects from other disease-driving mutations can be treated with DNMT inhibitors like 5-azacytidine or guadecitabine/decitabine (Issa et al. 2015; Gardin and Dombret 2017). (B) Drugs to target histone-modifying enzymes (writers+erasers) as well as readers (e.g., BET inhibitors) that are critical for AML maintenance and present vulnerabilities of the disease. Specific LSD1 inhibitors include GSK-LSD1 and ORY-1001 (Mohammad et al. 2015; Maes et al. 2018). Although PRC2 is deleted in a subset of AML, other AML subtypes such as KMT2A-re (MLL-rearranged) leukemias depend on functional PRC2. UNC1999, a dual inhibitor of EZH1/2 EZH1/2, impaired the growth of KMT2A-re in preclinical studies (Xu et al. 2015). DOT1L inhibitors (e.g., EPZ-5676) were also developed to target KMT2A(MLL)-re AML (Chen et al. 2016). The BRD 2/3/4 inhibitor OTX015 induces apoptosis in a variety of AMLs (Coudé et al. 2015). Inhibition of PRMT1 and PRMT5 show an antileukemia effect in distinct AMLs (Shia et al. 2012; Tarighat et al. 2016; Fedoriw et al. 2019). (C) Scheme illustrating the disruption of multiple layers of the epigenome in TET2mut cells. Loss of TET2 facilitates recruitment of the H3K4me1/2 histone demethylase LSD1 that inactivates enhancers at target genes. In addition, loss of TET2 results in promoter methylation at target genes such as GATA2. (D) Concept of targeting cooperating layers of the epigenome at enhancers and promoters in TET2mut AML to reconstitute enhancer–promoter interactions. Removal of 5mC promoter methylation by 5Aza treatment combined with LSD1 inhibition (GSK-LSD1) facilitates interactions of the LSD1-occupied enhancer and its target promoter, resulting in up-regulation of target genes like GATA2.