Figure 4.
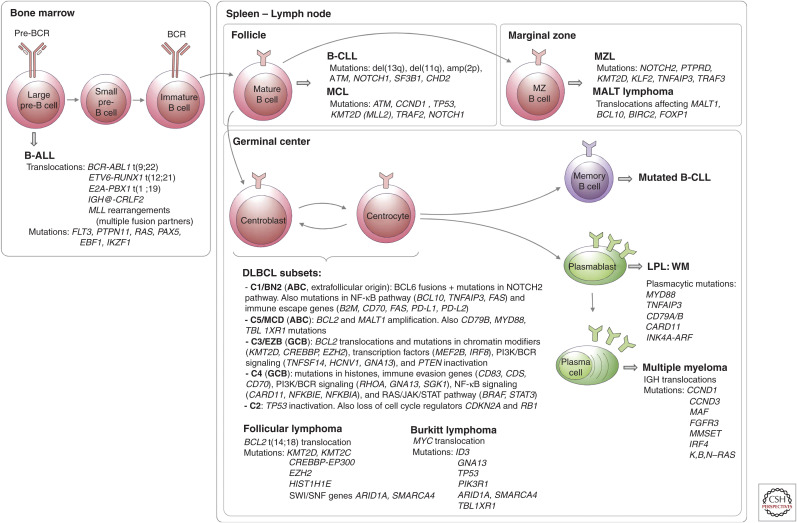
Frequent mutations during B-cell development. In the bone marrow, rearrangements of the immunoglobulin genes of B-cell precursors to form a B-cell receptor (BCR) generate DNA breaks that are occasionally resolved aberrantly, leading to chromosomal translocations (Fugmann et al. 2000). These are the most common genetic alterations in B-precursor acute lymphoblastic leukemia (B-ALL) (Mullighan 2012). The most frequent mature B-cell neoplasms that have their origin outside the germinal center (GC) are B-cell chronic lymphocytic leukemia (B-CLL), mantle cell lymphoma (MCL), marginal zone lymphoma (MZL), and mucosa-associated lymphoid tissue (MALT) lymphoma. B-CLL and MCL differ in their molecular pathways, genomic alterations, and clinical behavior, being more aggressive in naive-like- than memory-like-derived tumors. The pathogenesis of the two malignancies involves the BCR signaling, tumor cell microenvironment interactions, genomic alterations, and epigenome modifications (Zhang et al. 2014; Landau et al. 2015). MALT lymphoma is the commonest MZL type and presents recurrent chromosomal translocations, which usually lead to activation of the NF-κB pathway. Nodal and splenic MZLs share recurrent mutations affecting the Notch pathway and the transcription factor KLF2, but differ for the inactivation of two tumor-suppressor genes, detected exclusively (PTPRD) or much more commonly (KMT2D/MLL2) in the nodal type (Rossi et al. 2012; Spina et al. 2016). The presence of immunoglobulin mutations is evidence that the cell of origin of the tumor passed through the GC microenvironment. Follicular lymphomas, Burkitt lymphomas, and DLBCLs express GC B cell signature genes. In the GC, two molecular processes remodel DNA: immunoglobulin class switch recombination (CSR) and somatic hypermutation (SHM), mechanisms that predispose to chromosomal translocations and mutations (Muramatsu et al. 2000). DLBCL is a clinically and genetically heterogeneous disease and accounts for 35% of non-Hodgkin lymphomas. Based on transcriptional profiles, DLBCL is further classified into activated B-cell (ABC) and germinal center B-cell (GCB) subtypes (Alizadeh et al. 2000; Rosenwald et al. 2002). ABC-DLBCLs derive from B cells that are committed to plasmablastic differentiation (Victora et al. 2012). These tumors have increased NF-κB activity, genetic alterations in NF-κB modifiers and components of the BCR pathway, and perturbed terminal B-cell differentiation (Lenz et al. 2008; Ngo et al. 2011). GCB-DLBCLs originate from light-zone GC B cells (Alizadeh et al. 2000; Victora et al. 2012). These tumors have frequent alterations in chromatin-modifying enzymes, PI3 K signaling, and genetic alterations of BCL2 (Pfeifer et al. 2013; Basso and Dalla-Favera 2015). Modifications in these pathways could favor epigenetic reprogramming and escape from cellular immunity. Recent genomic profiles have identified sub-ABC and GCB-DLBCL clusters: C1-C5 in one study (of which two are GCB-, two are ABC-subtypes, and the fifth is mostly characterized by genomic instability and TP53 mutations) (Chapuy et al. 2018), BN2, MCD, N1 (mostly ABC), and EZB (mostly GCB) in a different study (Schmitz et al. 2018). Follicular lymphoma (FL) is characterized by a unique histology in which tumor B cells form follicle-like structures with large numbers of nonmalignant immune cells infiltrating within the follicular and interfollicular regions (Kridel et al. 2012). The most frequent genetic event is the t(14;18) translocation that places BCL2 under control of the immunoglobulin heavy-chain enhancer, which occurs in 90% of FL patients. Mutations in epigenetic modifiers (KMT2D, CREBBP, and EZH2) are also a hallmark of FL (Green 2018). These mutations result in altering normal B-cell differentiation programs and impeding GC exit (Green et al. 2015). Burkitt lymphoma is characterized by deregulation of the MYC gene through its translocation to one of the immunoglobulin loci (Love et al. 2012). LPL, lymphoplasmacytic lymphoma; WM, Waldenstrom macroglobulinemia.