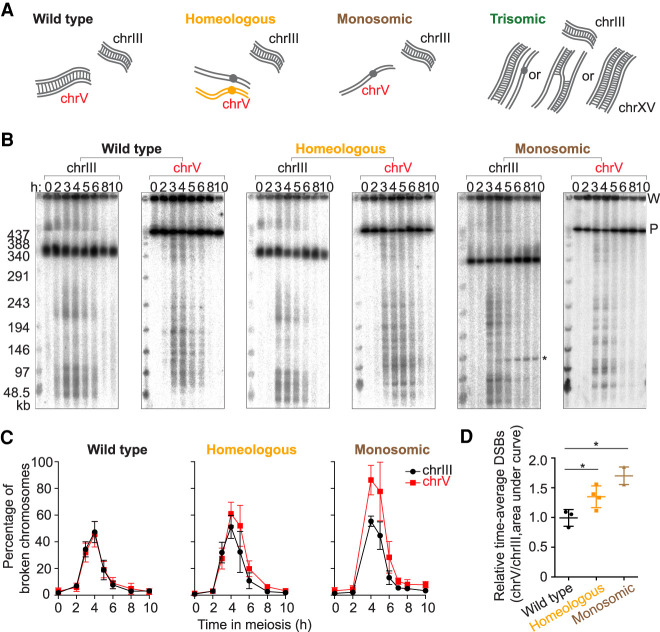
Figure 1.
Higher DSB levels on homolog engagement-defective chromosomes. (A) Cartoons of wild-type, homeologous, monosomic, and trisomic chromosome configurations. Gray lines are S. cerevisiae chromosomes and orange lines are S. pastorianus. The homeologous chrV pair rarely synapses or recombines. The trisomic chromosomes can adopt different synaptic configurations (”II + I,” partner switch, and triple synapsis). (B) Representative PFGE Southern blots probed for chrIII and chrV. P, signal from parental-length DNA; W, signal in wells. Asterisk indicates an ectopic recombination product between leu2::hisG (on chrIII) and ho::hisG (on chrIV) in the monosomic strain. The other strains do not form this product because they do not have a hisG insert at ho. (C) Poisson-corrected DSB quantification of PFGE Southern blots. (D) Quantification of time-averaged DSBs on chrV relative to chrIII for wild-type, homeologous, and monosomic strains. (*) P < 0.05, unpaired t-test. Error bars in C and D are mean ± SD except for the monosomic strain (mean ± range).