Figure 2.
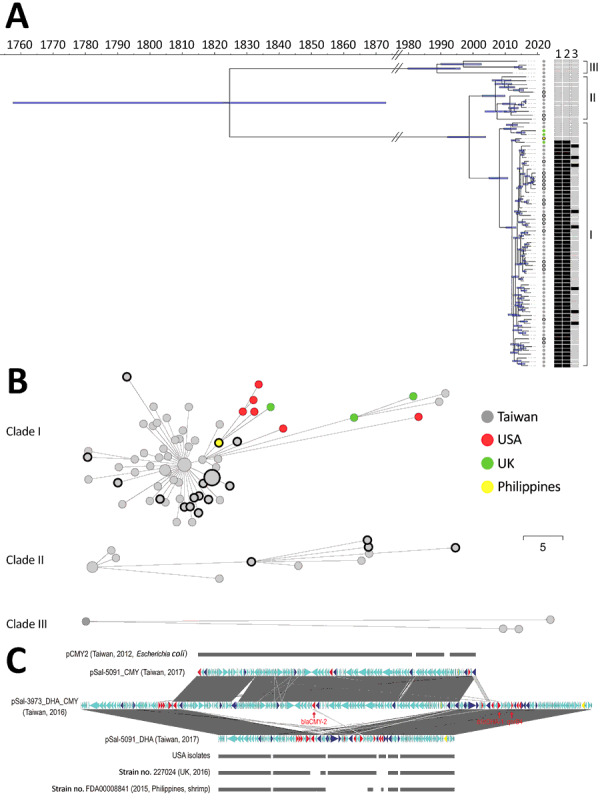
Genomic analysis of the outbreak caused by Salmonella enterica serotype Anatum, Taiwan. A) Dated phylogeny for Salmonella Anatum clinical isolates and food and environmental isolates. All isolates were divided into 3 clades, shown at right. The nodes’ colors represent the geo source; nodes with black rings were from meat or the environment, and the remainder were derived from the patients. The right heatmap represents the presence (in black) or absence (in gray) of key antimicrobial-resistance genes (1, blaDHA-1; 2, qnrB4; 3, blaCMY-2). B) Minimal spanning tree based on alleles identified through core genome multilocus sequence typing. Dots with black circles represent food isolates; the others are clinical isolates. The collection date for the 6 US isolates in panel B was missing in GenBank and therefore not included in panel A. Scale bar indicates 5 single nucleotide polymorphisms. C) Gene structure of multidrug-resistant plasmids in Salmonella Anatum in Taiwan compared with international isolates. Two types of plasmids were identified in the clade I Salmonella Anatum isolates in Taiwan. One carried blaCMY-2, with its structure being shown by pSal-5091_CMY. A similar plasmid, pCMY2 (GenBank accession no. LC019731.1), is shown. The other carried blaDHA-1; its structure is shown by pSal-5091_DHA. International isolates shown in the figure, whose genomes also were downloaded from GenBank (Appendix), possess very similar plasmids. In certain isolates, the 2 plasmids can integrate into 1 large plasmid, with its structure shown by pSal-3973_DHA_CMY. Red genes represent antimicrobial-resistance genes; blue genes represent transposase/integrase genes; and yellow genes represent Inc-determinant genes.
