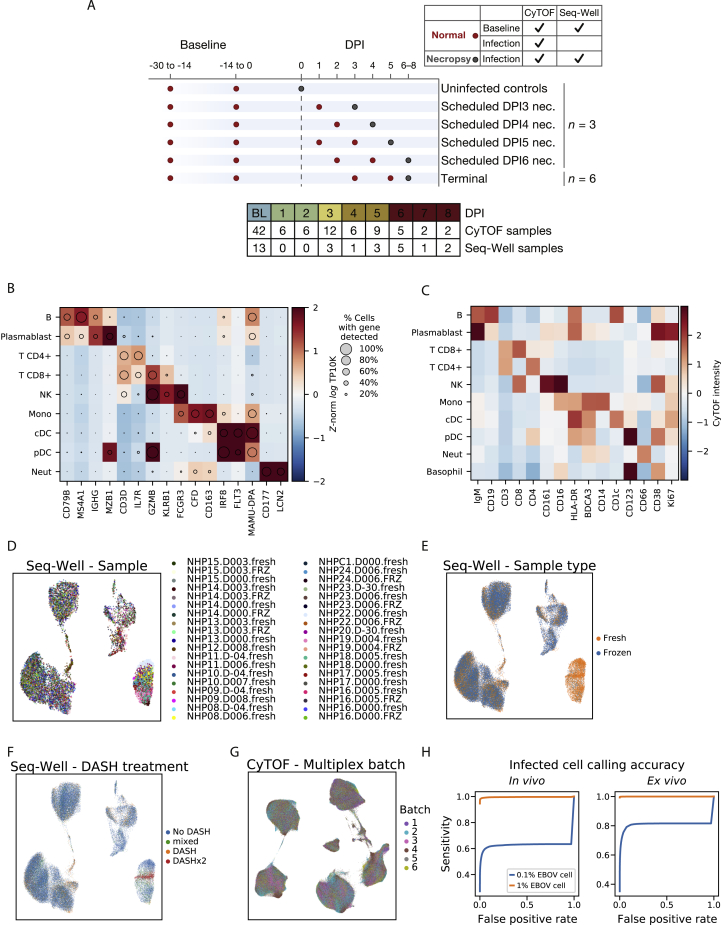
Figure S1.
Cell-Type Markers for Seq-Well and CyTOF Clusters, Related to Figure 2
(A) Overview of study cohorts and blood draw timelines. Animals were grouped into cohorts with pre-scheduled necropsy times (at baseline, or day post infection [DPI] 3, 4, 5, 6 - n = 3 each), or allowed to progress until clinical score exceeded 10 (terminal), predetermined euthanasia criteria. Dots: scheduled blood draws for each cohort; red: intermediate (non-necropsy) draw; gray: draw that coincided with euthanasia and necropsy. Necropsy and baseline normal draws were used for Seq-Well and CyTOF, while intermediate post-infection draws were available only for CyTOF.
(B) Expression profiles of cell-type marker genes (columns) for cell-type clusters (rows) based on the in vivo Seq-Well data. Circle area represents the percentage of cells in each group in which the gene was detected, and color denotes the average expression level (loge TP10K).
(C) Average expression (Z-normalized CyTOF intensity) profiles of cell-type marker genes (columns), for cell-type clusters (rows), based on the CyTOF data.
(D) Uniform Manifold Approximation and Projection (UMAP) embedding of post-integration Seq-Well data, colored by the sample source (NHP, DPI, and whether the sample was loaded for Seq-Well without any freezing [.fresh] or was frozen with cryoprotectant and thawed prior to Seq-Well [.FRZ]). A maximum of 500 cells per sample is plotted to increase representation across samples.
(E) UMAP embedding of Seq-Well data, colored by whether cells were processed fresh (orange) or after freeze/thaw (blue) prior to Seq-Well.
(F) UMAP embedding of Seq-Well data, colored by depletion of abundant sequences by hybridization (DASH) treatment. We developed a DASH-based method to remove a PCR adaptor artifact from some Seq-Well sequencing libraries (STAR Methods), and performed this 0 times (No DASH, blue), 1 time (DASH, orange), or 2 times sequentially (DASHx2, red). For a few samples, we sequenced ‘No DASH’ and ‘DASH’ libraries and merged the reads (mixed, green).
(G) UMAP embedding of batch-corrected CyTOF data, colored by the multiplex batch in which it was pooled and analyzed by CyTOF.
(H) Receiver operating characteristic curves for identifying EBOV-infected cells. Estimates of sensitivity to detect an infected cell at various false positive rate thresholds in vivo (left) and ex vivo (right). Curves are estimated separately for a hypothetical viral load of 0.1% (blue line) and 1% (orange line).