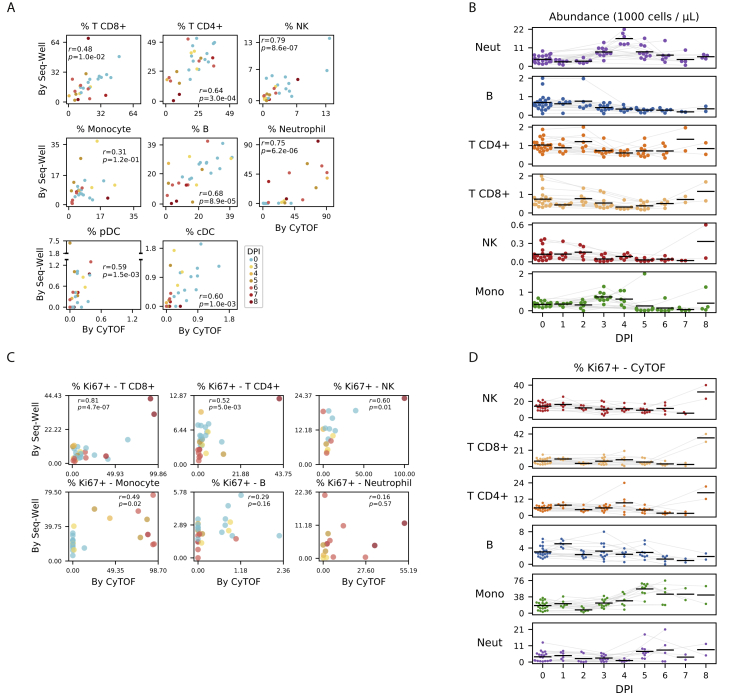
Figure S3.
Estimates of Cell-Type Abundance and Proliferation over the Time Course, Related to Figure 2
(A) Scatterplot of the percentage of cells of each cell type in a sample, inferred from CyTOF (x axis) or Seq-Well (y axis), for several cell types (panels). Each dot represents a sample colored by DPI. Pearson correlation coefficients (r) and p-value are provided.
(B) Estimates of the abundance of each cell type (rows) for each NHP (individual markers) in units of 1000 cells per μL of whole blood, based on integration of CyTOF and complete blood count (CBC) information. Black line: mean value of each DPI; gray lines: serial samples from the same NHP.
(C) Scatterplots of the percentage of Ki67-positive cells in a sample inferred from CyTOF (x axis) or Seq-Well (y axis) for several cell types (panels). Each dot represents a sample colored by DPI. Cells with smoothed expression of MKI67 (the gene coding for Ki67) > 0.1 are called Ki67-positive by Seq-Well. Cells with CyTOF intensity > 1.8 are called Ki67-positive by CyTOF.
(D) Estimates of the percentage of Ki67-positive cells (CyTOF intensity > 1.8) of each cell type (rows) for each animal replicate (markers). Black line: mean value of each DPI; gray lines: serial samples from the same NHP.