Figure S4.
Quantification of Cytokine Expression and Enrichment of Response Signatures, Related to Figures 3 and 4
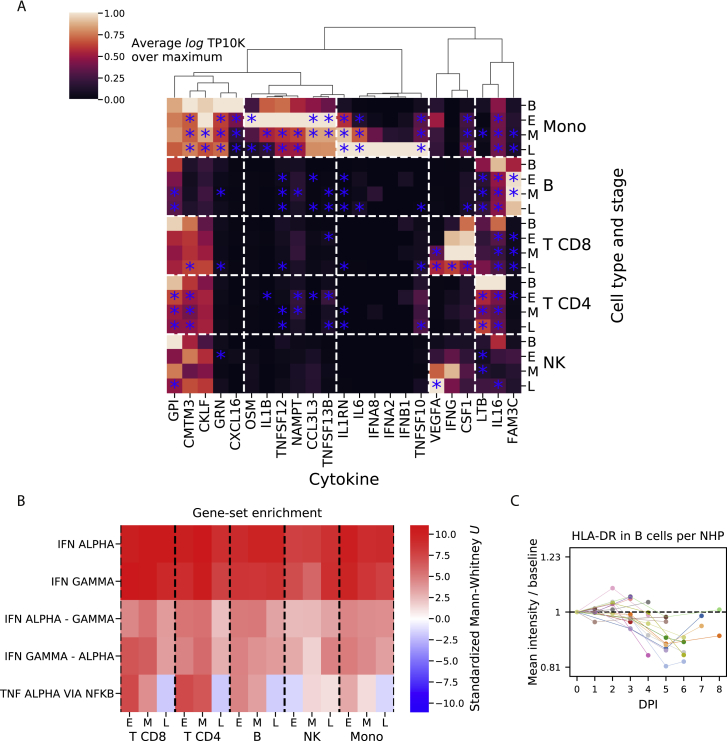
(A) Average expression values (loge TP10K) of literature-annotated cytokines (columns) across cell types and stages of acute EVD (rows). Values are plotted as a ratio relative to the maximum across cell types and stages. Values that are statistically different from baseline (p < 0.05) are indicated with a blue star.
(B) Heatmap of rank-sum test statistics for comparison of differential expression log fold-changes of genes in a gene set (rows) compared to genes not in the set. The log fold-changes were defined from differential expression profiles of each cell type at each EVD stage (columns) relative to baseline. Five gene sets were tested — three from the Hallmark database (IFN ALPHA, IFN GAMMA, and TNF ALPHA VIA NFKB) (Liberzon et al., 2015) and 2 constructed from the hallmark sets, as uniquely IFNα-regulated genes in “IFN ALPHA” but not “IFN GAMMA” (“IFN ALPHA - GAMMA”), and vice versa for uniquely IFNγ-regulated (“IFN GAMMA - ALPHA”). See also Table S3.
(C) Fold change (log2 scale) in average HLA-DR CyTOF intensity on B cells at each DPI relative to baseline for each PBMC sample. Colored lines connect serial samples from the same NHP.