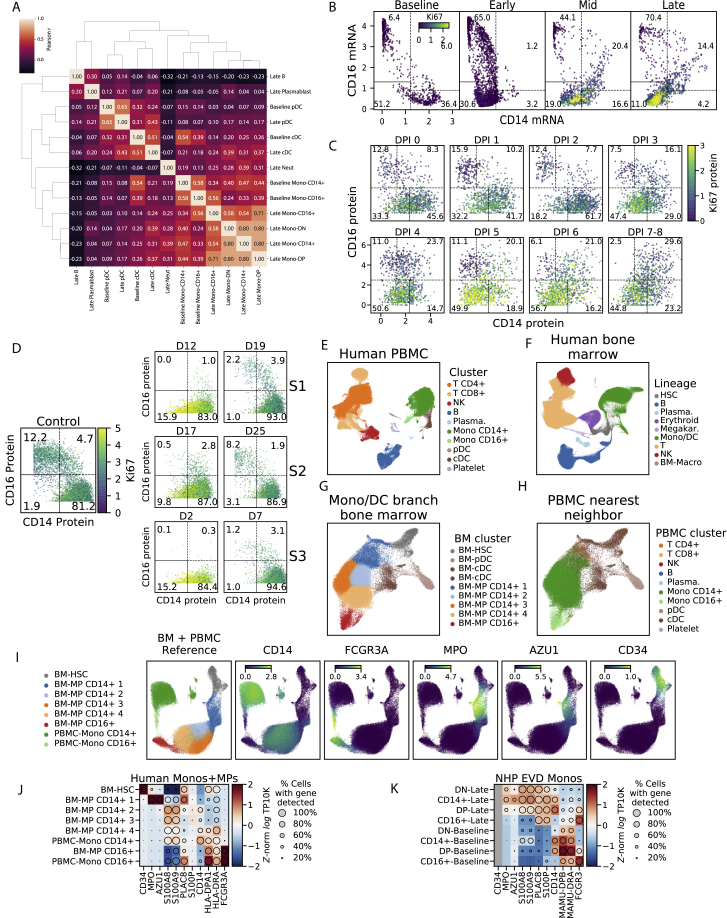
Figure S5.
Extended Characterization of Interferon and Double-Negative CD14– CD16– Monocytes, Related to Figure 5
(A) Clustermap of pairwise Pearson correlations between cell type clusters at baseline and late EVD. Correlations are computed on average loge TP10K expression values of overdispersed genes. DN and DP monocytes at late EVD are more similar to monocytes (including baseline CD14+s) than other cell types.
(B) Scatterplot of MAGIC-smoothed expression values (loge TP10K) of CD14 and CD16 for monocytes in baseline, early, mid, and late disease stages. Cells are colored by smoothed expression levels of MKI67 (the gene coding for Ki67 protein). Boxes: CD14+, CD16+, DN, and DP subsets described in the text; numbers: percentage of cells falling into each subset.
(C) Scatterplot of protein expression (CyTOF intensity) of CD14 and CD16 for 1,000 randomly sampled monocytes at each DPI. Cells are colored by Ki67 expression. Boxes: CD14+, CD16+, DN, and DP subsets described in the text; numbers: percentage of cells falling into each subset.
(D) Scatterplot of protein expression (CyTOF intensity) of CD14 and CD16 for monocytes during human EVD. Left: monocytes from healthy human controls. Right: monocytes from 3 EVD cases (S1, S2, and S3) at various days post symptom onset. Cells are colored by Ki67 marker intensity. Boxes: CD14+, CD16+, DN, and DP subsets described in the text; numbers: percentage of cells falling into each subset.
(E) UMAP embedding of healthy human PBMCs dataset, colored by annotated cluster assignment, based on known marker genes. (Plasma.: Plasmablast).
(F) UMAP embedding of healthy bone marrow cells, colored by cluster assignment, based on marker genes. (HSC: hematopoietic stem cell, Plasma.: Plasmablast, Megakar.: Megakaryocyte, Mono/DC: monocyte and dendritic cell, BM-Macro: bone marrow macrophage).
(G) UMAP embedding of sub-clustered HSC and monocyte/dendritic lineage cells. (BM: bone marrow, MP: monocyte progenitor)
(H) Same UMAP embedding as Figure S5G, but colored by the cluster identity of their nearest neighbor in the human PBMC dataset (Figure S5E).
(I) UMAP embedding of the merged reference dataset of healthy bone marrow HSCs and monocyte lineage cells and PBMCs. Left sub-panel is colored by cluster assignment. Right sub-panels are colored by marker gene expression (loge TP10K).
(J) Expression profiles of selected genes for human bone marrow monocyte progenitors (BM-MPs) and human circulating monocytes (PBMC-Monos). Circle area: percentage of cells in which the gene was detected; color: average expression (Z-normalized loge TP10K).
(K) Expression profiles of selected genes for NHP monocyte subsets at baseline or late EVD for orthologs of the genes in (J). Circle area: percentage of cells in which the gene was detected; color: average expression level (Z-normalized loge TP10K). CD34 is grayed out because it is detected in <10 cells.