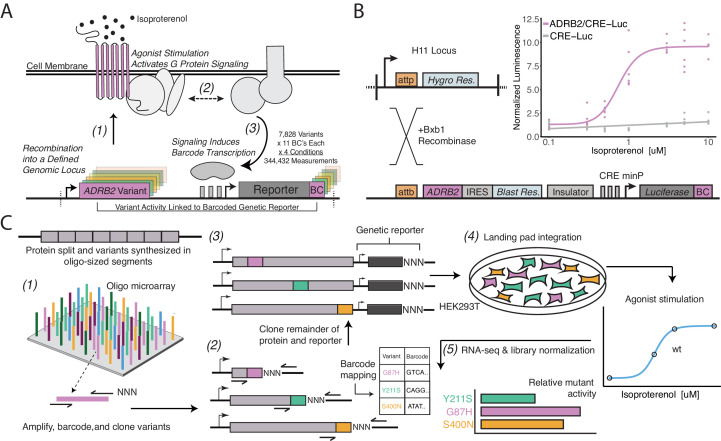
Figure 1. A platform for deep mutational scanning of GPCRs.
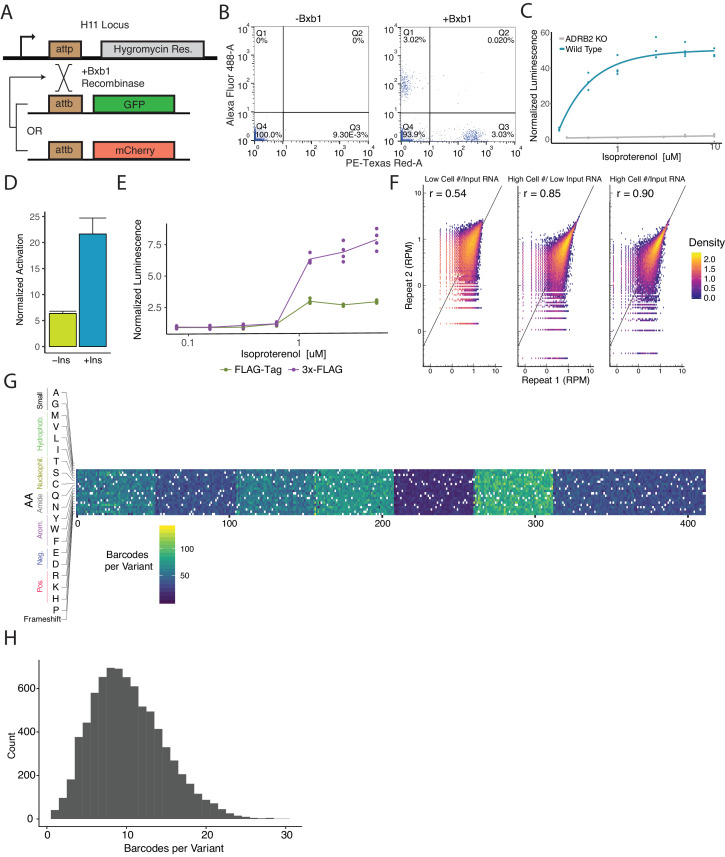
(A) Overview of the multiplexed GPCR activity assay. Plasmids encoding ADRB2 variants, a transcriptional CRE reporter of signaling activity, and 15 nucleotide barcode sequences that identify the variant are integrated into a defined genomic locus such that one variant is present per cell. Upon stimulation by isoproterenol, G-protein signaling induces transcription of the CRE genetic reporter and the barcode. Thus, the activity of a given variant is proportional to the amount of barcode mRNA which can be read out in multiplex by RNA-seq. (B) Schematic detailing the recombination of the reporter-receptor expression plasmid into the landing pad locus. Top right: activation of the CRE reporter integrated with (purple) or without (grey) exogenous ADRB2 into the landing pad when stimulated with isoproterenol in ΔADRB2 cells via a luciferase CRE reporter gene assay. (C) Overview of library generation and functional assay. Missense variants are synthesized on an oligonucleotide microarray, the oligos are amplified with random DNA barcode sequences appended, and the variants are cloned into wild-type background vectors. Barcode-variant pairs are mapped with next-generation sequencing and the remaining wild-type receptor and CRE reporter sequences are cloned into the vector. Next, the variant library is integrated en masse into the serine recombinase (Bxb1) landing pad engineered at the H11 locus of ΔADRB2 HEK293T cells. This integration strategy ensures a single pair of receptor variant and barcoded CRE reporter is integrated per cell and avoids crosstalk. After selection, the library is stimulated with various concentrations of the β2AR agonist, isoproterenol. Finally, mutant activity is determined by measuring the relative abundance of each variant’s barcoded reporter transcript with RNA-seq.