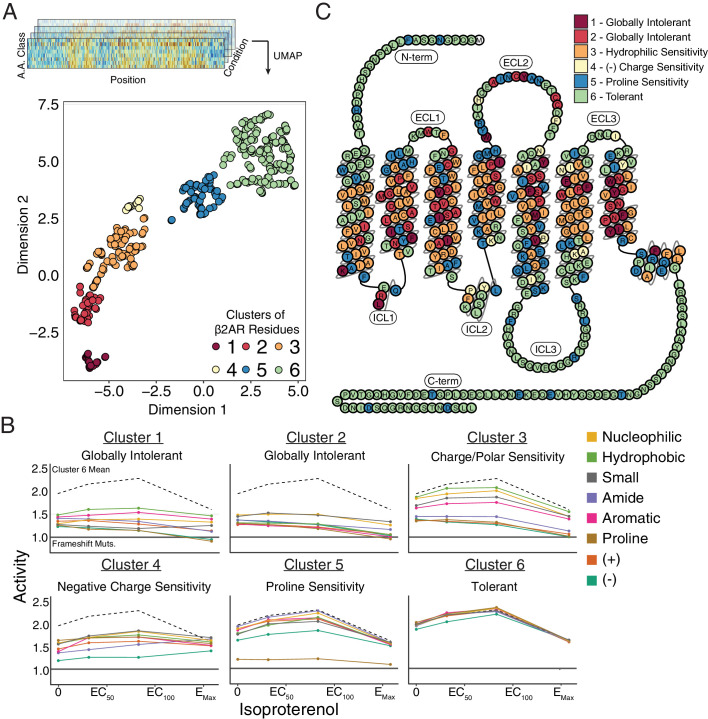
Figure 4. Unsupervised learning segregates residues into clusters with distinct responses to mutation.
(A) Amino acids were segregated into classes based on their physicochemical properties and mean activity scores were reported by class for each residue. With Uniform Manifold Approximation and Projection (UMAP) a 2D representation of every residue’s response to each mutation class across agonist conditions was learned. Each residue is assigned into one of six clusters using HDBSCAN (see Figure 4—figure supplement 1). (B) Class averages for each of these cluster reveal distinct responses to mutation. The upper dashed line represents the mean activity of Cluster 6 and the lower solid line represents the mean activity of frameshift mutations. (C) A 2D snake plot representation of β2AR secondary structure with each residue colored by cluster identity.