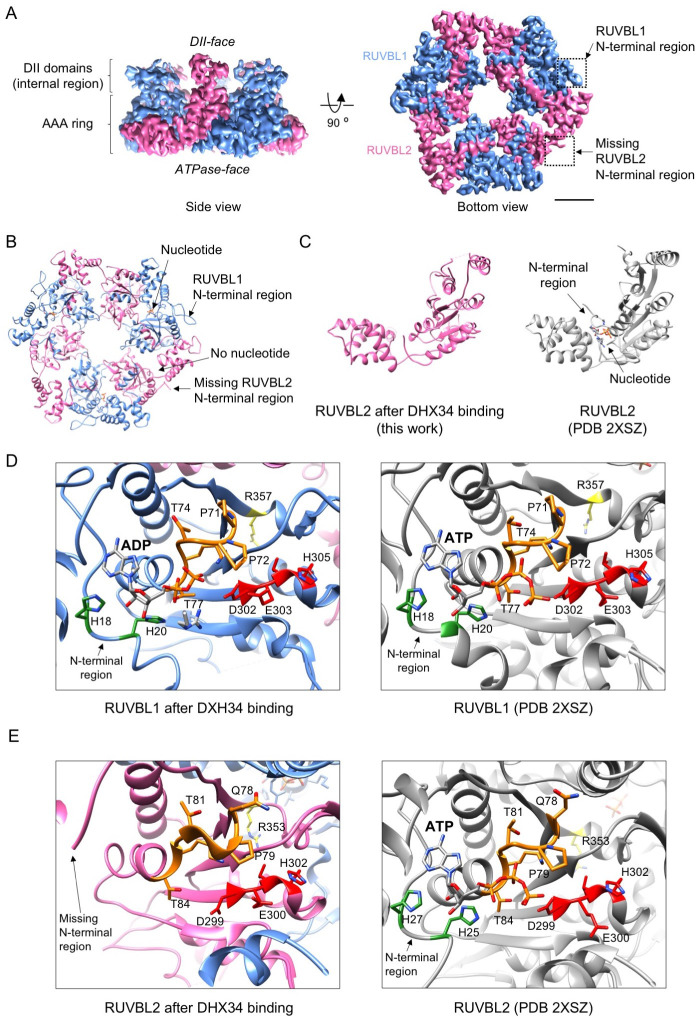
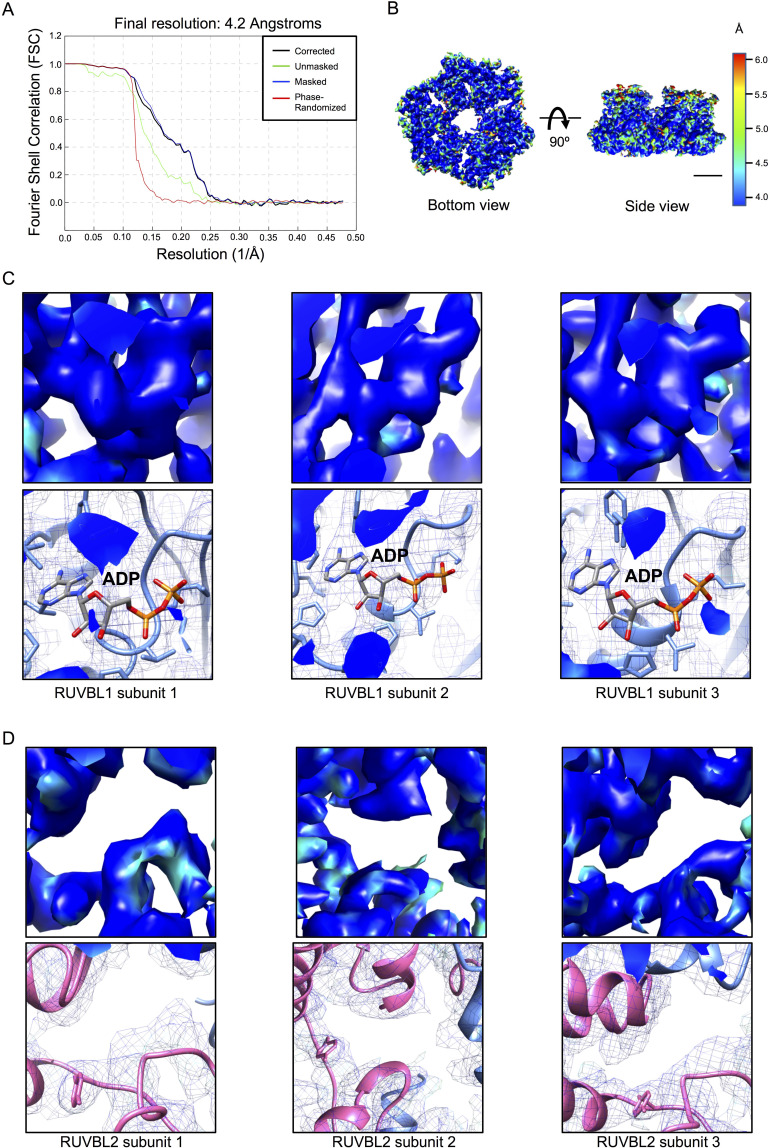
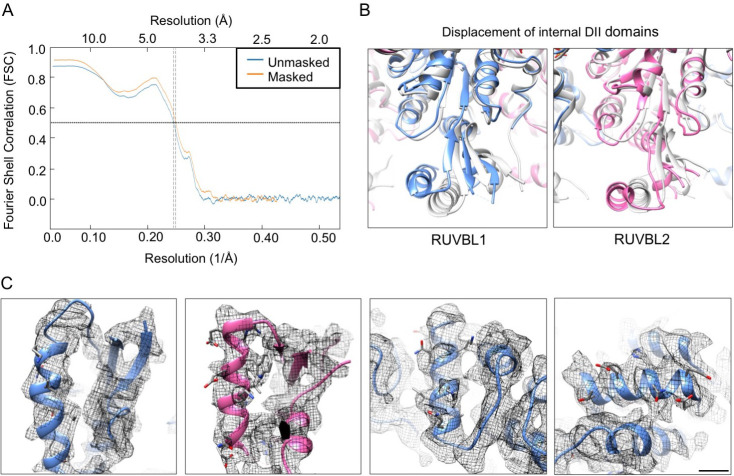
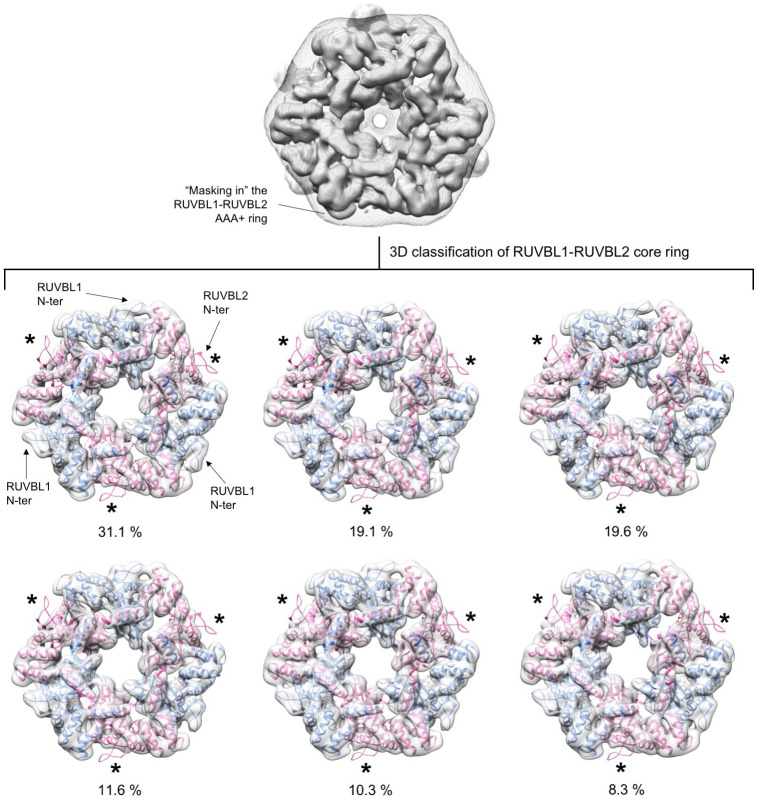
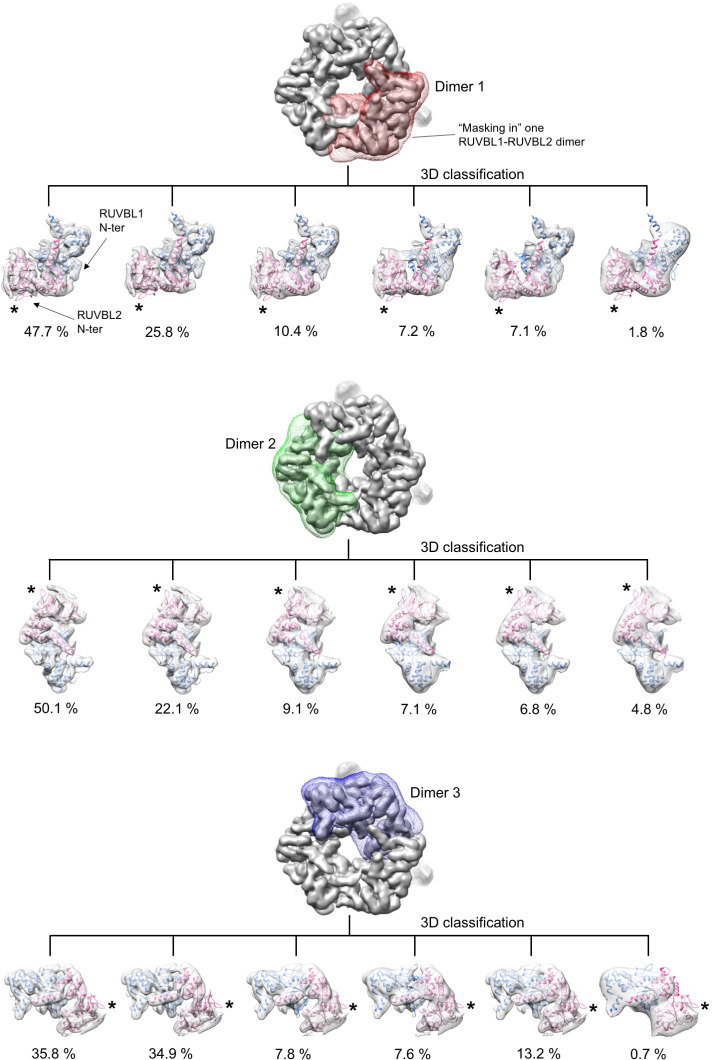
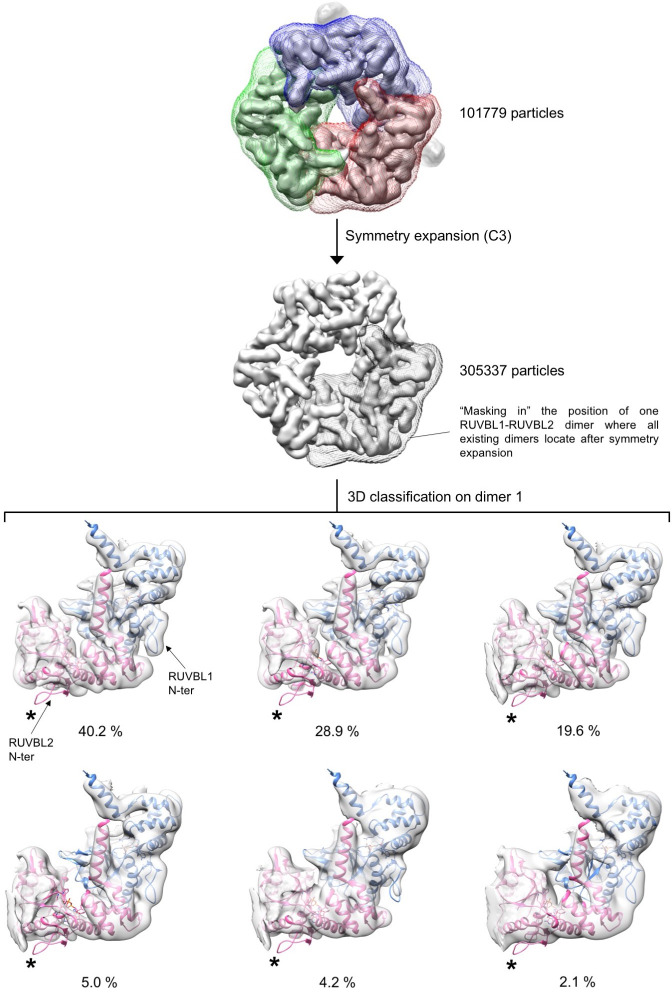
Figure 3. DHX34 induces large conformational changes in RUVBL2.
(A) Side and bottom views of the RUVBL1-RUVBL2 ring obtained after refinement without the influence of DHX34 and the OB-fold domains. Squares highlight N-terminal segments of RUVBL1 (blue) and RUVBL2 (pink). Scale bar represents 25 Å. The presence and absence of RUVBL1 and RUVBL2 N-terminal regions is indicated only in one copy of each subunit, but it applied to all the subunits in the complex. (B) Bottom view of the atomic structure of RUVBL1-RUVBL2 ring modeled from the cryo-EM density. Color codes are as in (A). (C) Right panel: a view of the nucleotide binding region in RUVBL2 from the crystal structure of RUVBL1-RUVBL2 (PDB 2SXZ) in gray color; left panel: similar view of RUVBL2 in RUVBL1-RUVBL2 after DHX34 binding (this work, pink). (D) Close-up view of the nucleotide-binding regions in RUVBL1, comparing the structure after DHX34 binding (left panel) and the crystal structure of the RUVBL1-RUVBL2 complex (PDB 2SXZ) (right panel) in gray. N-terminal histidines (H18 and H20) are indicated in gray, Walker A residues in orange, Walker B in red, and the Arg finger in yellow. (E) As in (D) but for the RUVBL2 subunit. Color codes for relevant and catalytic motifs are represented as in (D).