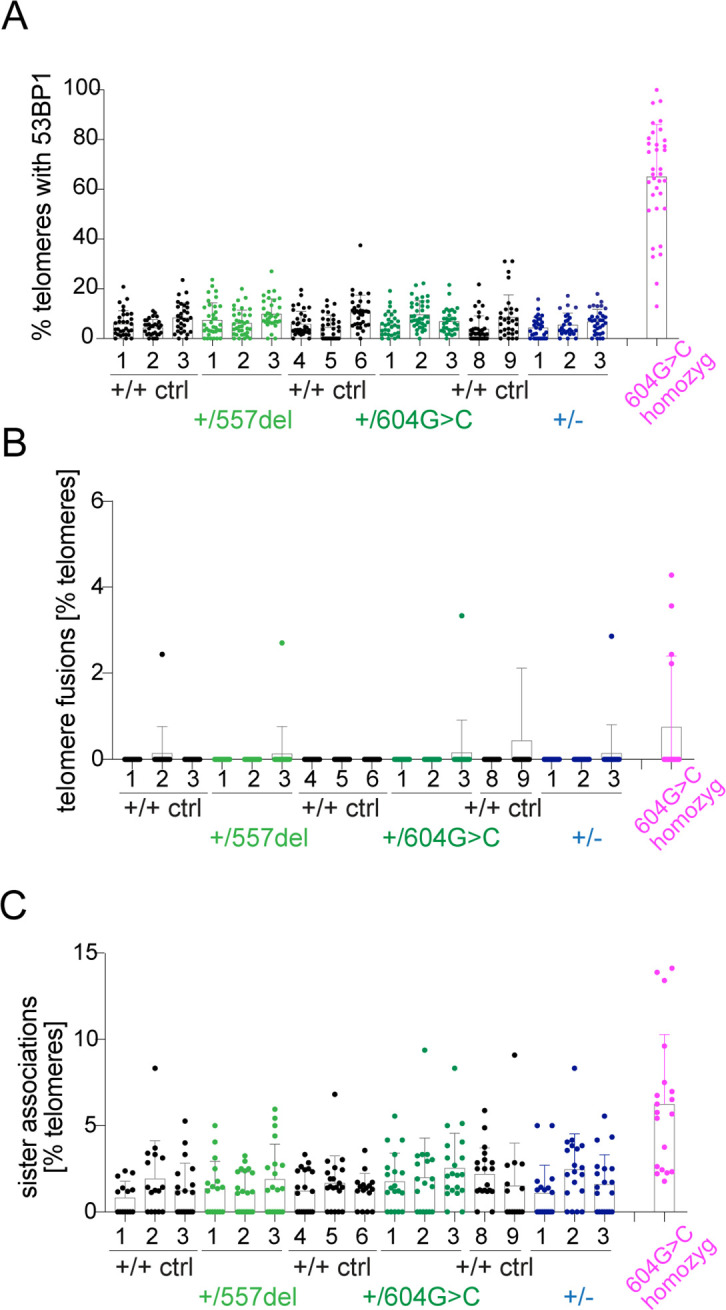
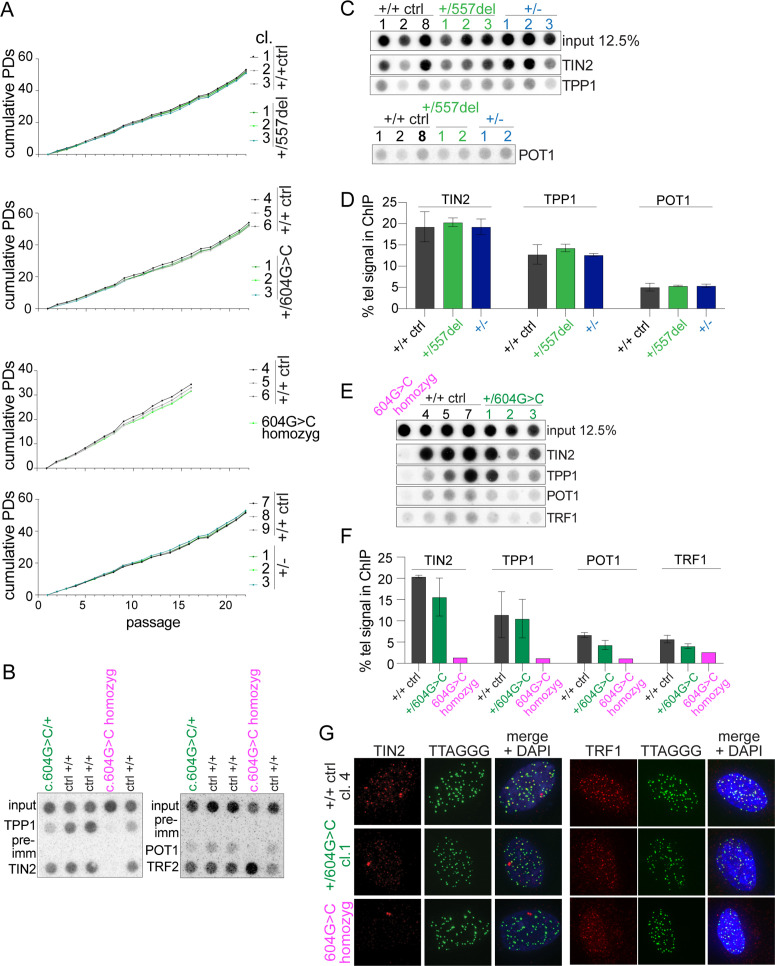
Figure 4. Heterozygous TINF2 mutations do not cause telomere damage or genome instability.
(A) Immunoblot for TIN2 and γtubulin in control cells and the indicated clones with targeted TINF2 alleles. (B) Quantification of the immunoblot shown in A. Unpaired t-test was used to determine significance. Symbols: *p<0.05; ns, not significant (0.16). (C) Representative images of TIF analysis in control and indicated TINF2 mutant cells. IF for 53BP1 (red), telomeric FISH (green) and DNA (DAPI, blue). (D) Quantification of percentage of telomeres colocalizing with 53BP1 foci. Data from ≥50 nuclei per cell line, with three cell lines per genotype (with the exception of the single c.604G > C homozyg clone). (E) Representative metaphase spreads of cells with mutated TINF2 alleles. Sister telomere associations (>), telomere fusions (*), and a marker chromosome found in all clones (marker) are indicated. Telomere FISH (red), centromere FISH (green) and DNA (DAPI, gray). (F) Quantification of telomere fusions ≥20 spreads per cell line, with three cell lines per genotype (except for the single 604G > C homozyg clone). (G) Quantification of the % of telomeres found in sister associations. Data from ≥20 spreads per cell line; three cell lines per condition, except for the single 604G > C homozyg clone. For the quantification in (B), (D), (F), and (G) means ± SD and individual data points are shown. One-way ANOVA with Tukey post-test was used to determine significance, p-values: ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05. ns, not significant. See also Figure 4—figure supplements 1–6.
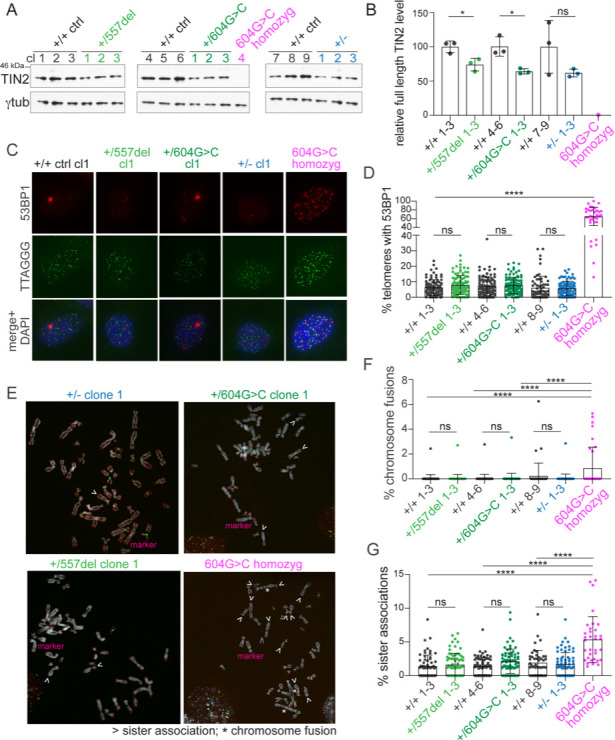
Figure 4—figure supplement 1. Transcript analysis in 604G > C/+ cells reveals presence of two alternative TINF2 transcripts (604G > C I, 604G > C II).
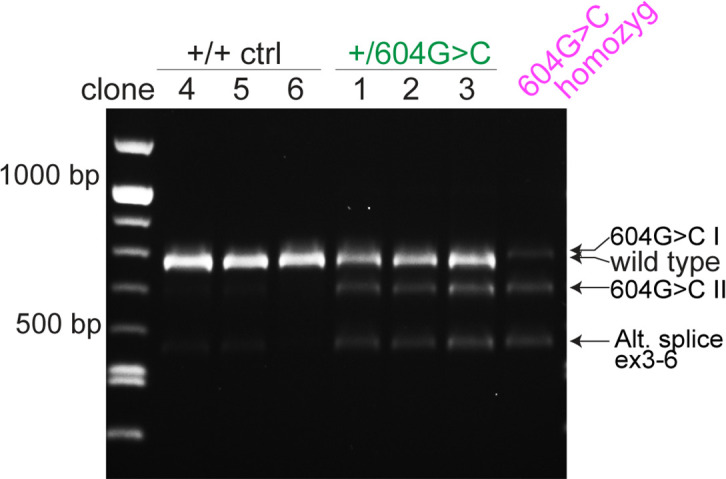
Figure 4—figure supplement 2. Knock-in strategy for introduction of c.557del and c.604G > C mutations into RPE1 cells.
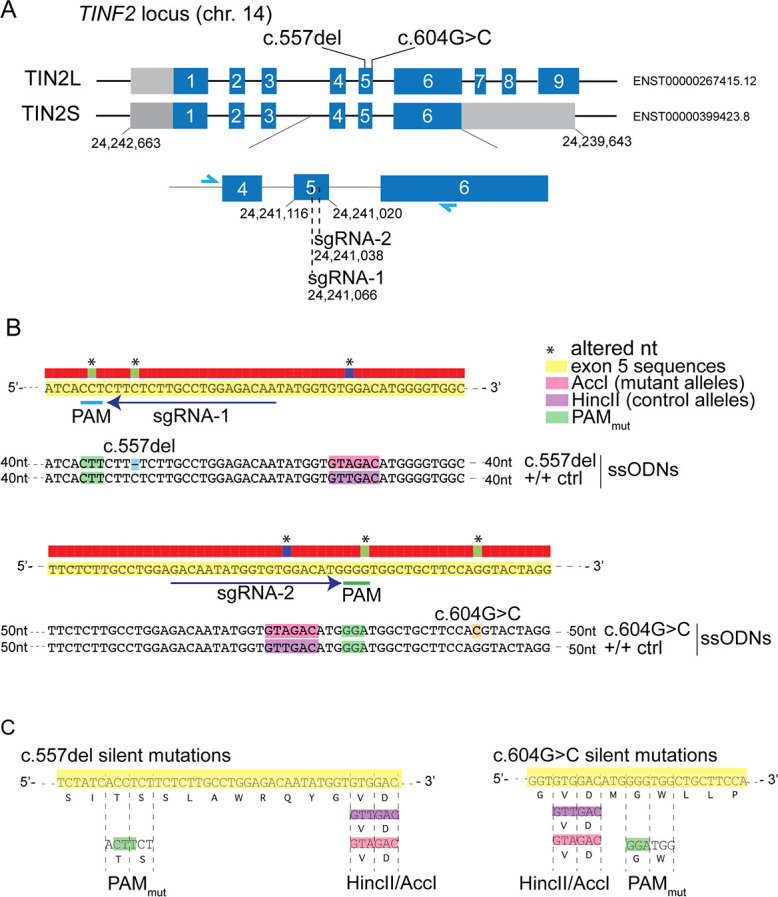
Figure 4—figure supplement 3. Strategy to generate TIN2+/- RPE1 clones.
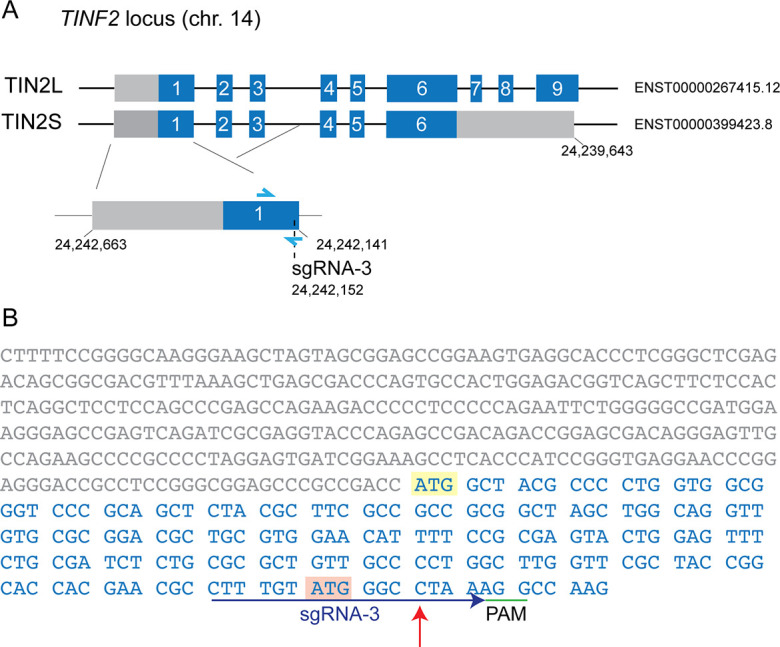
Figure 4—figure supplement 4. Sanger sequencing of CRISPR/Cas9-engineered clones with TINF2 mutations.
Figure 4—figure supplement 5. Characterization of cells with targeted TINF2 alleles.
Figure 4—figure supplement 6. Representation of TIFs, telomere fusions, and sister associations in the individual cell lines.