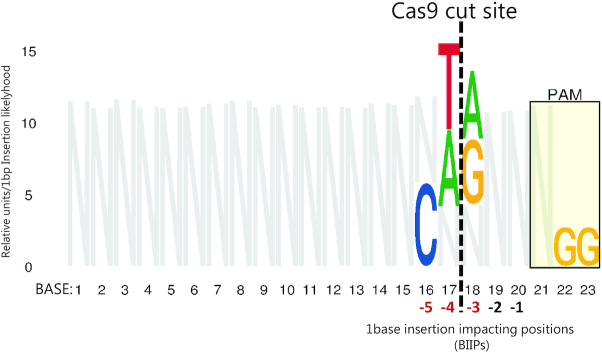
Figure 9.
IDAA based identification of 1-bp insertion impacting positions (BIIPs) after SpCas9:gRNA editing. Starting with the indel summary data generated from >800 individually validated gRNA design IDAA profiles (GlycoCRISPRcollection Addgene https://www.addgene.org/browse/article/28192658/), a BBAM model (BennettBrodyAnalysis Model) was trained for each type of indel that occurred more than 30 times as a sign of a significant repair outcome. From these models, the 1-bp insertion prediction BBAM model performed the best and was further investigated. The 1-bp insertion rate was calculated for each base in each position across the protospacer (number of 1-bp insertions/number of non-1-bp insertions). From this, outliers were defined as those bases where the +1 insertion rate had a modified Z-score greater than 3.0. Following that, we let the value of N at each base (except for the last two bases) be the sum of the value of the bases not determined to be an outlier. Only significant BIIPs are shown in color and all other sequence positions with N (in gray).