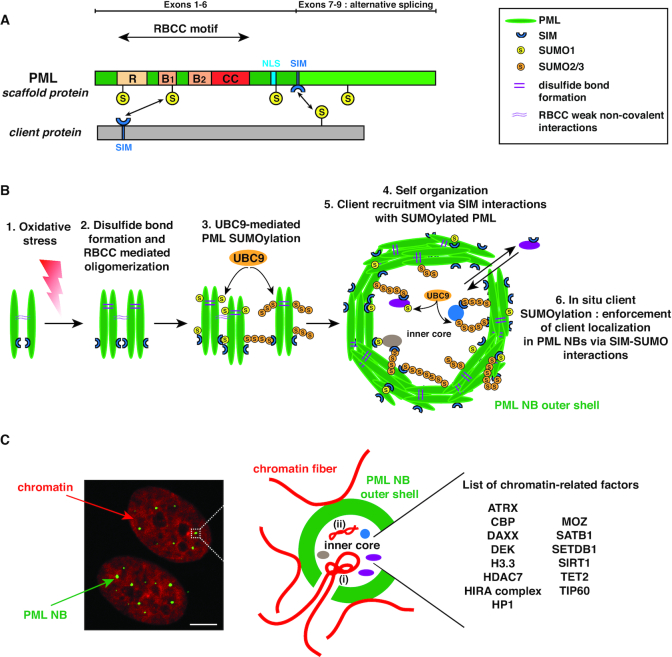
Figure 1.
Structure of PML and organization of PML NBs. (A) Structure of the PML protein scaffold. All PML isoforms (I–VII), ranging from 882aa (PML-I) to 435aa (PML-VII), possess a conserved RBCC/TRIM motif in their N-terminal part. The different C-terminal parts of PML-I to VI are generated through alternative splicing of the 3′ exons 7 to 9 of the unique PML gene, while PML-VII only possesses exons (1–4 and 7b). SUMO modification sites (S) are indicated at lysine positions K65, K160, K490 and K616. The NLS (Nuclear Localisation Sequence) and the SIM (SUMO Interacting Motif) are indicated. NB: PML structure is not to scale. (B) Formation of canonical PML NBs. (1) Oxidative stress triggers PML cross-linking by disulfide bond formation. (2) Together with RBCC weak non-covalent interactions, this triggers oligomerization of non SUMOylated PML proteins. (3) UBC9-mediated (poly-)SUMOylation of PML then allows multiple SUMO-SIM interactions, (4) which stabilize the formation of the self-organized matrix-associated outer shell, possibly involving liquid-liquid phase separation mechanisms. Of note SUMO1 modification (yellow) is mostly present in the PML NB outer shell, while the poly-SUMO2/3 chains (orange) present in the shell also protrude to variable degrees in the interior of the PML NB. (5) Client proteins are recruited in the outer shell (eg Sp100 not shown) as well as in the inner core through specific interactions of their SIM with the SUMOylated-PML scaffold. (6) UBC9 SUMOylation of client proteins then enforces their sequestration in PML NBs. Turnover of client proteins is relatively rapid ranging from seconds to a few minutes. (C) PML NBs are interspersed in the chromatin. (left) Immunofluorescence analysis of human primary BJ fibroblasts stained by PML (green) and DAPI (red). Scale bar is 10μm. (right) Scheme showing PML NBs (green) surrounded by chromatin loops (red). Cellular loci, such as telomeres, can localize partly within PML NBs in specific cases (i) (see main text). Chromatin-related factors (histone modifiers, histone readers and histone chaperones) as well as viral genomes (ii) localize inside PML NBs.