Figure 4.
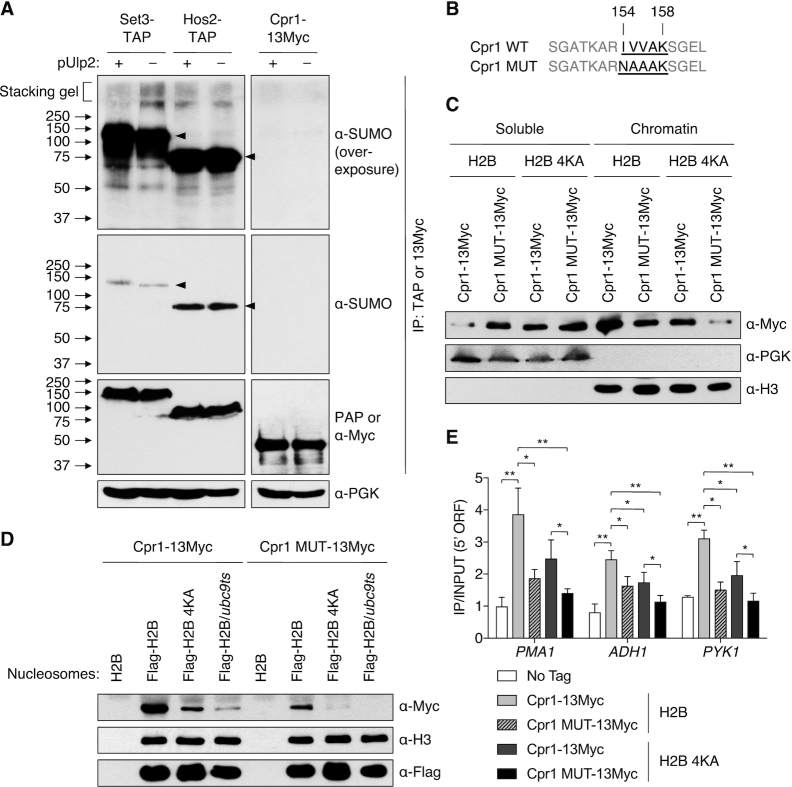
Mutation of the SET3C Cpr1 subunit SIM blocks chromatin binding. (A) IP of Set3-TAP and Hos2-TAP with IgG-Sepharose and Cpr1–13Myc with anti-Myc agarose from denatured yeast extracts in ulp2Δ strains with or without YCplac33-ULP2. Precipitated proteins were followed by immunoblotting. Anti-PGK was used as a loading control. The top panels show overexposure of SUMO blot. Potentially polySUMO-conjugated Set3-TAP is seen in the stacker in lane 2. Arrowheads, unmodified TAP-tagged proteins. (B) Mutation of SIM within Cpr1. Putative SIM residues 154–158 are bolded and underlined. (C) Chromatin association assays using htb1–1 htb2–1 cpr1Δ strains with pRS314 plasmids expressing Flag-H2B or Flag-H2B-4KA and pRS425-GPD-Cpr1–13Myc or pRS425-GPD-Cpr1 MUT-13Myc. Chromatin and soluble fractions were analyzed by immunoblotting. (D) In vitro nucleosome binding assays of Cpr1 WT and MUT. Nucleosomes isolated on anti-Flag agarose from the indicated strains were incubated with extracts from cpr1Δ cells (MHY10887) carrying pRS425-GPD-borne Cpr1–13Myc or Cpr1-MUT-13Myc, precipitated, and then immunoblotting with the indicated antibodies. (E) ChIP assays at the indicated 5′ ORF regions using strains in (C). An untagged strain (MHY10244) was used as a negative control. Data represented mean ± SD of triplicates. *P <0.05; **P <0.01 (Student's t test between indicated pairs of values).