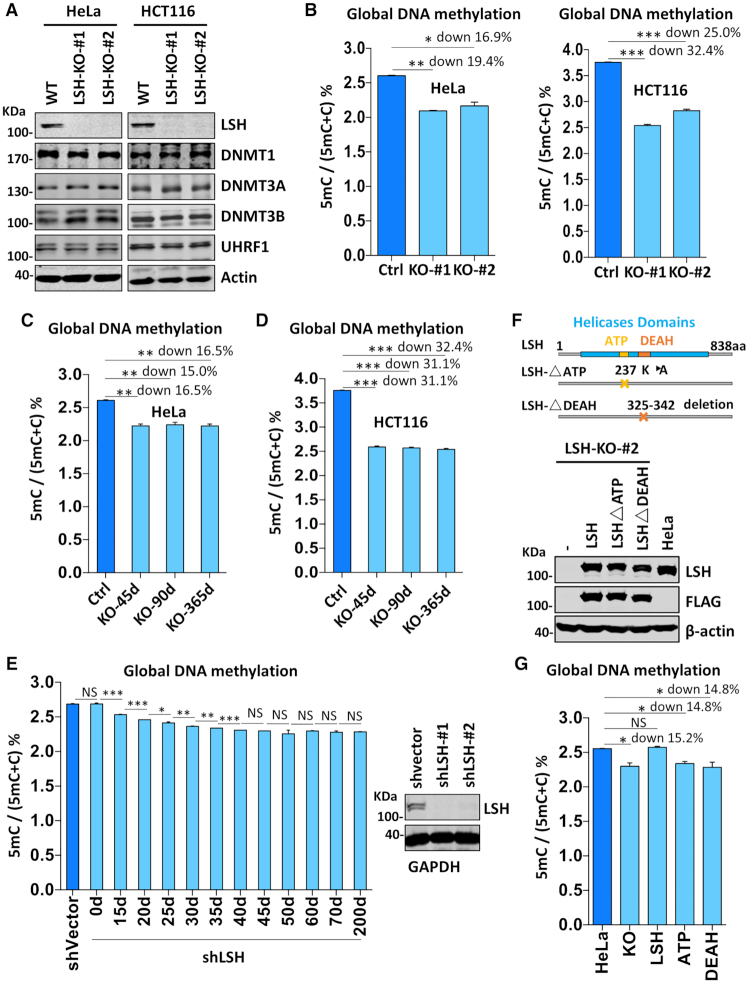
Figure 1.
LSH is required for DNA methylation in various cells. (A) Western blots showing that knockout of LSH did not affect the levels of DNMT1, DNMT3A, DNMT3B and UHRF1 proteins in HeLa and HCT116 cells. (B) Quantification of the levels of 5mC in genomic DNA derived from control and LSH-KO cells by HPLC. The levels of 5mC were shown as 5mC/(5mC + C)%. *P < 0.05; **P< 0, 005, ***P< 0. 0005, n = 3, error bar represents SEM in all 5mC measurement. (C) The levels of 5mC in genomic DNA derived from HeLa LSH-KO#2 cells consecutively cultured for various days determined by HPLC. The culture was regularly splited every three days. ** P< 0.005, n = 3, error bar represents SEM in all 5mC measurement. (D) The levels of 5mC in genomic DNA derived from HCT116 LSH-KO#1 cells consecutively cultured for various days determined by HPLC. ***P< 0. 0005, n = 3, error bar represents SEM in all 5mC measurement. (E) The levels of 5mC in genomic DNA derived from LSH-knockdown HeLa cells cultured for various times determined by HPLC. *P < 0.05; ** P< 0.005, ***P< 0. 0005, n = 3, error bar represents SEM in all 5mC measurement. (F) Western blots showing re-expression of wild-type and mutant LSH in stable culture derived from HeLa LSH-KO#2. Also shown at top are schematic drawings of wild-type and mutant LSH. (G) DNA methylation measurement by HPLC showing re-expression of wild-type but not mutant LSH restored DNA methylation in HeLa LSH-KO#2 cells. *P < 0.05, n = 3, error bar represents SEM in all 5mC measurement.