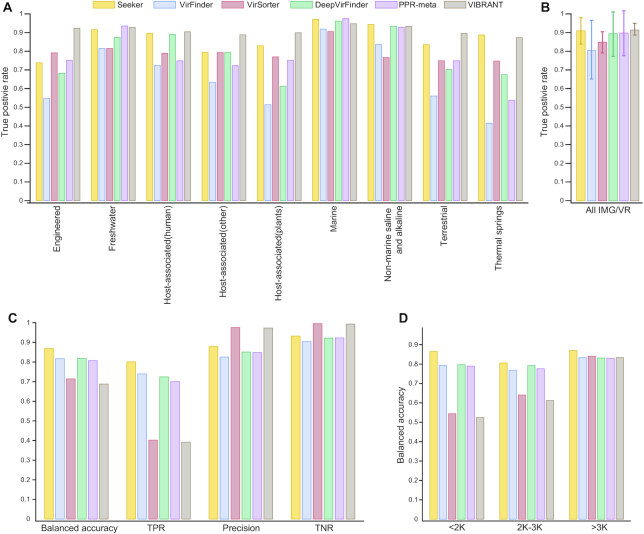
Figure 2.
Comparison of the performance of Seeker to those of other approaches for the IMG/VR dataset and for short phage and bacterial sequences. (A) True positive rates of Seeker (yellow), VirFinder (Blue), VirSorter (red), DeepVirFinder (green), PPR-Meta (purple) and VIBRANT (brown) for phages in the IMG/VR data set. (B) Overall true positive rates of the nine data points in panel (A); the error bars show standard deviations of the rates; the color code is the same as in panel A. (C) Balanced accuracy, true positive rate (TPR), precision (positive predictive rate) and true negative rate (TNR) of the six methods, for short sequences mimicking metagenomics data; the color code is the same as in panel A. (D) Balanced accuracy of the six methods for short sequences mimicking metagenome projects, for three ranges of sequence lengths; the color code is the same as in panel A.