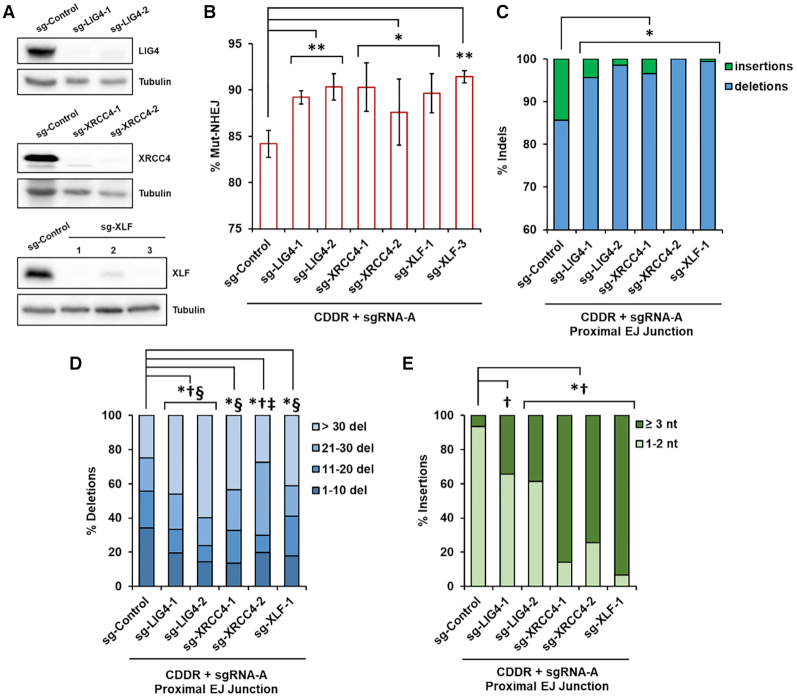
Figure 5.
The cNHEJ proteins DNA Ligase IV, XRCC4, and XLF suppress mutagenic NHEJ and deletion mutagenesis in the repair of DSBs. (A) Immunoblots of protein lysates from independent clones of U2OS-CDDR-Clone-1 cells deleted of LIG4, XRCC4 or XLF. (B) Percentage of cells with single DSBs repaired via Mut-NHEJ (% mCherry-ve cells) in the indicated cell lines. Data represent the mean of three independent experiments ± s.d. Statistical significance was determined using two-tailed Student's t-test; *P< 0.05, **P< 0.01. (C) Frequency of deep sequencing reads categorized as insertions or deletions among total indels at repair junctions following the induction of single DSBs within the CDDR reporter. Statistical significance was assessed using chi-squared tests with Yates' continuity correction. Bonferroni correction and an additional stringency of >1.5-fold change was applied to all comparisons; P< 0.0001 and >1.5-fold change: *insertions. (D) Size distribution among deletions at repair junctions following the induction of single DSBs within the CDDR reporter. Statistical significance was assessed using chi-squared tests with Yates' continuity correction and Bonferroni correction; P< 0.0001 and >1.5-fold change: *1–10 del, †11–20 del, ‡21–30 del, §>30 del. (E) Frequency of insertions of either 1–2 nt or ≥3 nt at repair junctions following the induction of single DSBs within the CDDR reporter. Statistical significance was assessed using chi-squared tests with Yates' continuity correction and Bonferroni correction; P< 0.0001 and >1.5-fold change: *1–2 nt ins, †≥ 3 nt ins.