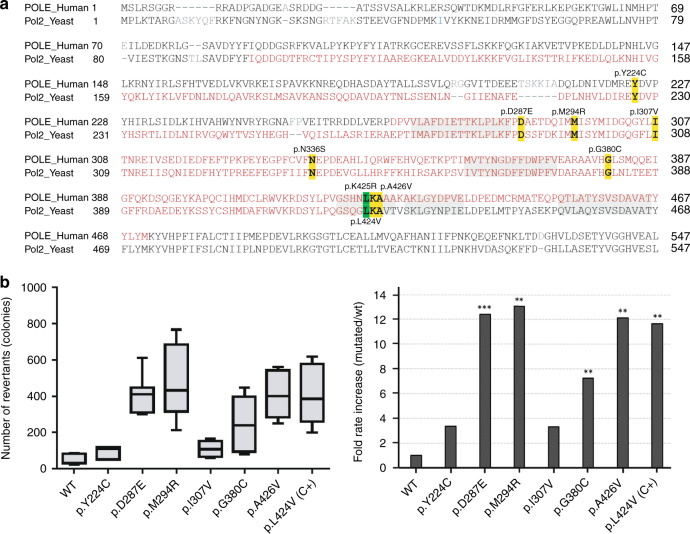
Fig. 2. Functional analysis carried out in Schizosaccharomyces pombe for variants located within (or close to) the POLE exonuclease domain (ED).
(a) Alignment of human POLE and their homolog in yeast (Pol2). The identified variants are highlighted in yellow (conserved residues) and the POLE p.Leu424Val, used as positive control, in green. ED is depicted in red (human: residues 268–471, yeast: residues 98–428) and its sequence motifs44 are shaded in gray. (b) Left panel: box plots showing mutation rates of ade6-485 S. pombe (number of colonies) for pol2 wildtype (WT, negative control), ED mutation-positive control (pol2-Leu425Val; C+) and the corresponding variants. Right panel: fold rate increase relative to the median number of revertants in the WT. Data obtained from two independent experiments performed in triplicate. *p value = 0.01, **p value = 0.01–0.001, and ***p value < 0.001 indicate the differences with the WT clone, and were calculated using the Mann–Whitney nonparametric test.