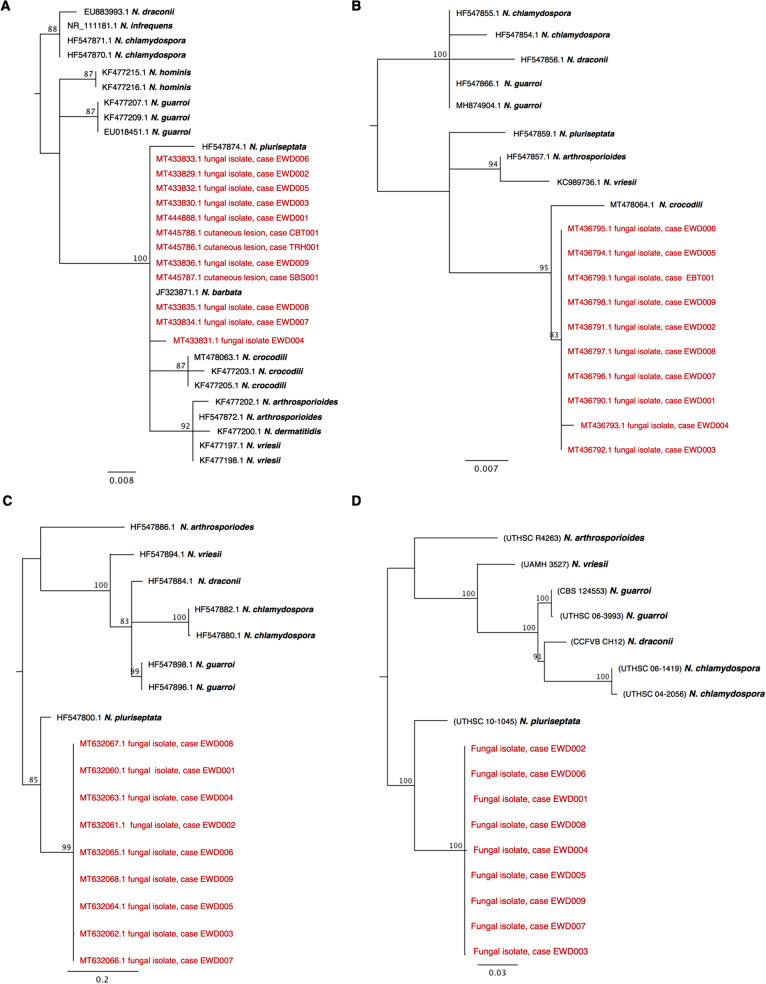
Figure 4.
Maximum likelihood trees depicting the relationship of fungal isolates obtained in this study with other members of the Nannizziopsis genus for which similar sequences were available. Analyses were performed using (A) ITS2, (B) 28S, (C) β-tubulin, and (D) concatenated (ITS + 28S + β-tubulin) sequence alignments. Midpoint-rooted consensus trees were constructed from 750 original trees produced using PhyML, based on a generalized time reversible substitution model, bootstrapped with 1000 random re-samplings of the data. Branches are supported by consensus support threshold values > 75%. Scale bars represent the mean number of nucleotide substitutions per length.