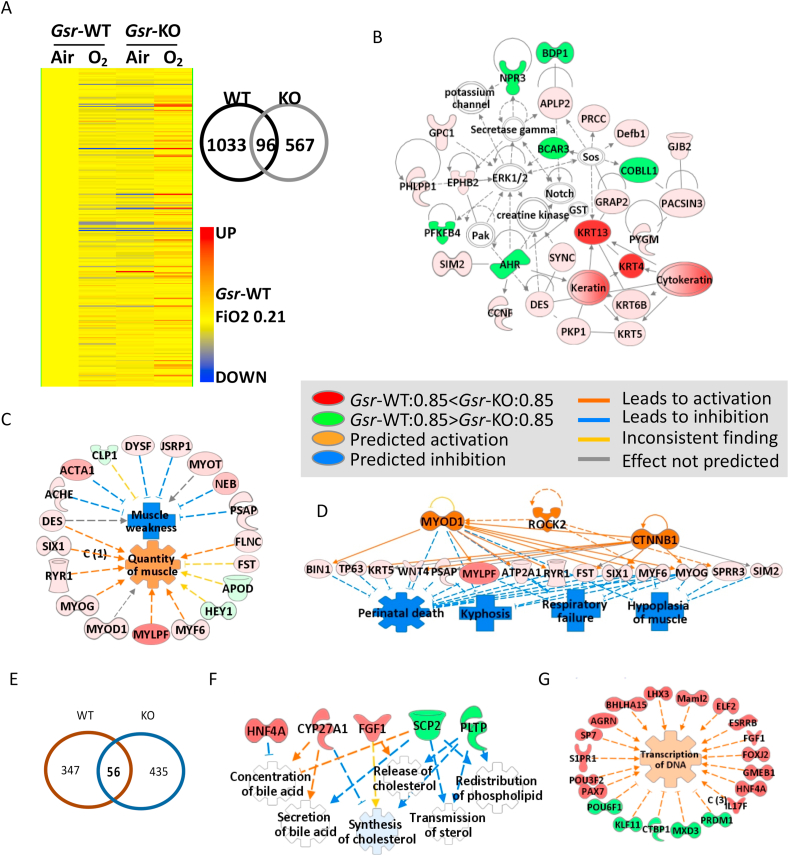
Fig. 7.
Effects of glutathione reductase (Gsr) deficiency and hyperoxia (85% O2) on lung transcriptomics. (A) Heat map from hierarchical clustering analysis depicts lung expression profiles of Gsr-dependently hyperoxia (0.85 FiO2)-responded genes at postnatal day 5 (PND5) after hyperoxia exposure (PND0-PND5, n = 303, 2-way ANOVA with p < 0.01). Color bar indicates average expression intensity (n = 3/group) normalized to wild-type (Gsr-WT)-Air (0.21 FiO2) group. Venn Diagram analysis depicted number of genotype-specific hyperoxia responsive genes determined by moderated t-test in Gsr-WT and Gsr-deficient (Gsr-KO) neonates. (B) Hyperoxia altered lung genes involved predominantly in cytoskeleton and skeletal/muscular development and tissue and cell morphology in Gsr-KO neonates. (C) The key molecular network of the Gsr-dependently altered neonatal lung genes by hyperoxia was cytoskeleton and skeletal-muscular function and tissue morphology and development. (D) Hyperoxia-altered genes in Gsr-KO neonates may inhibit respiratory failure and neonatal death through activation of upstream molecules such as myogenic differentiation 1 (Myod1) and β-catennin (CTNNB1). (E) Venn Diagram analysis depicted number of genotype-specific hyperoxia responsive genes following recovery from neonatal hyperoxia recovery within Gsr-WT and Gsr-KO adult mice. (F) Neonatally exposed hyperoxia was predicted to inhibit genes involved in lipid metabolism (e.g., bile acid, cholesterol) and enhance genes involved in transcription of adulthood lungs in Gsr-KO mice. Analysis was done by GeneSpring and Ingenuity Pathway Analysis software. Molecules colored by expression levels of Gsr-KO/0.85 FiO2 at PND5 (vs Gsr-WT/0.85 FiO2). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)