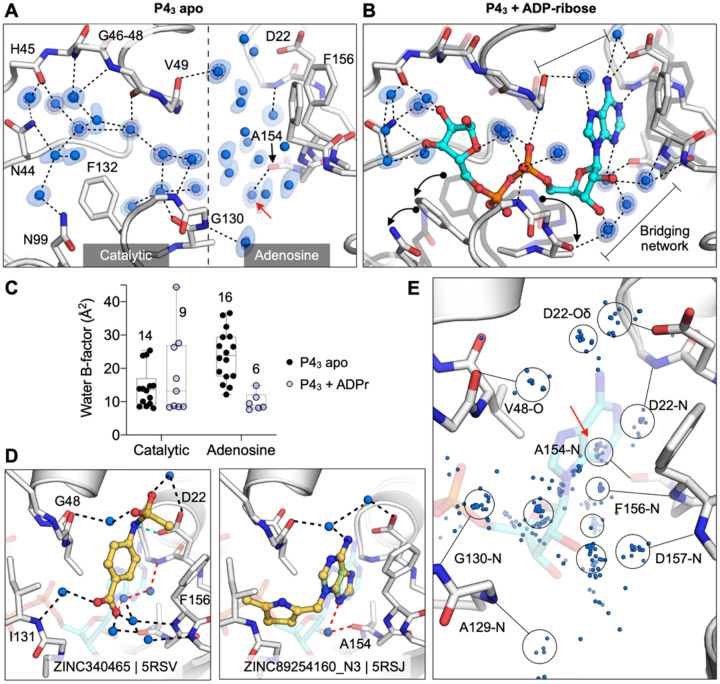
Figure 8. Water networks in the active site are displaced as well as used by fragments for bridging interactions.
A) Water networks in the apo enzyme (P43 crystal form). Waters are shown as blue spheres, with electron density contoured at 5 σ (blue mesh) and 1.5 σ (blue surface). Hydrogen bonds are shown as dashed lines (distances are 2.6–3 Å). B) Water networks in the Mac1-ADPr complex. ADP-ribose is shown as cyan sticks. Conformational changes upon ADP-ribose binding are highlighted with black arrows. C) Comparison of crystallographic B-factors of water molecules in the catalytic site and adenosine site. The range and 95% confidence interval are shown. D) Examples of the role of water networks in fragment binding. Right: ZINC340465 (PDB 5RSV) makes a single hydrogen bond to the protein (green dashed line), but makes five hydrogen bonds via water molecules. Although few fragments hydrogen bond directly to the backbone oxygen of Ala154 (Figure 4E), several fragments interact with this residue via bridging water molecules (red dashed line) including ZINC89254160_N3 (PDB 5RSJ). E) Plot showing all water molecules that lie within 3.5 Å of a non-carbon fragment atom. Water molecules are shown as blue spheres, with the major clusters circled. The cluster in a red circle bridges fragments and the Ala154 backbone oxygen.