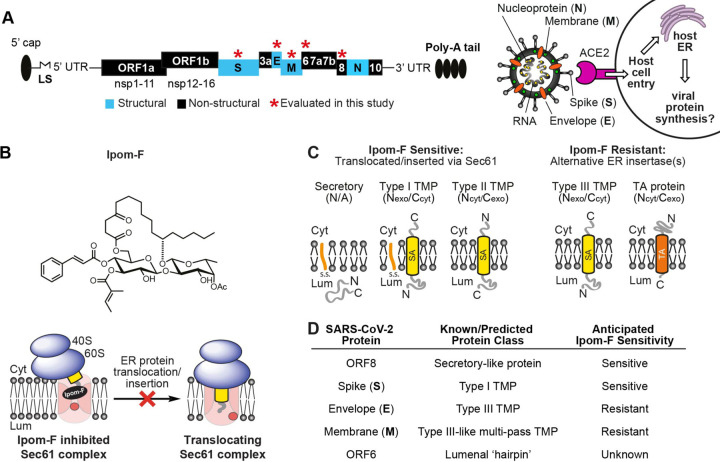
Fig. 1. Ipom-F as a potential inhibitor of SARS-CoV-2 viral protein synthesis.
(A) Schematic of (+) ssRNA genome architecture of SARS-CoV-2 (29903 nt) containing 5’ capped mRNA with a leader sequence (LS), 3’ end poly-A tail, 5’ and 3’ UTRs and open reading frames (ORFs): ORF1a, ORF1b, spike (S), ORF3a, envelope (E), membrane (M), ORF6, ORF7, ORF8, nucleoprotein (N) and ORF10 (Firth, 2020; Naqvi et al., 2020). An important mode of SARS-CoV-2 host entry proceeds via interaction of the viral S protein with human angiotensin-converting enzyme 2 (ACE2) (Walls et al., 2020). (B) Structure of Ipomoeassin-F (Ipom-F), a small molecule inhibitor of Sec61-mediated protein translocation. (C) Ipom-F efficiently blocks membrane translocation of secretory proteins and insertion of single-pass type I and type II TMPs, but not insertion of type III TMPs or tail-anchored (TA) proteins. SA denotes a signal anchor. (D) Based on known/predicted membrane topology of SARS-CoV-2 proteins, and sensitivity of comparable host cell proteins (Zong et al., 2019; O’Keefe et al., 2020 submitted), likely sensitivity to Ipom-F was anticipated.