| Protein sample |
5 mg ml−1 in 20 mM HEPES pH 7.4, 100 mM NaCl, 1 mM DTT |
7.5 mg ml−1 in 20 mM HEPES pH 7.4, 100 mM NaCl, 1 mM DTT |
| |
|
|
| Initial crystal |
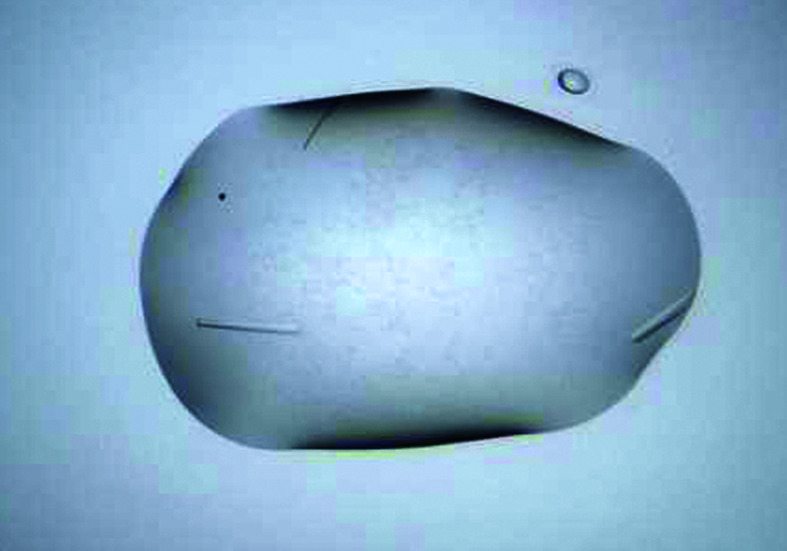
|
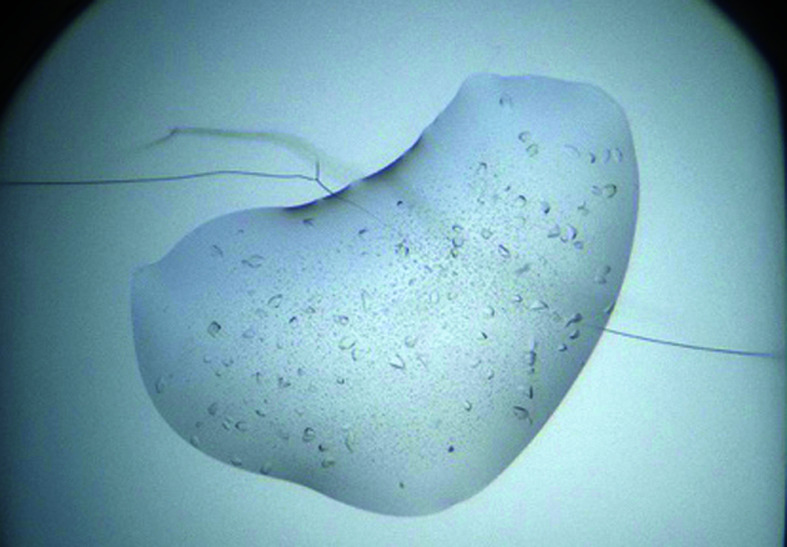
|
| Screen |
The PEGs II Suite |
The PEGs II Suite |
| Condition |
H9: 0.05 M magnesium acetate, 10%(w/v) PEG 8000, 0.1 M sodium acetate |
H8: 0.2 M calcium acetate, 10%(w/v) PEG 8000. 0.1 M HEPES pH 7.5 |
| Protein sample |
7 mg ml−1 in 20 mM HEPES pH 7.4, 100 mM NaCl, 1 mM DTT |
7 mg ml−1 in 20 mM HEPES pH 7.4, 100 mM NaCl, 1 mM DTT |
| Optimized condition |
0.05 M magnesium acetate, 16%(w/v) PEG 8000 |
0.2 M calcium acetate, 10%(w/v) PEG 8000, 0.1 M HEPES pH 7.5, 3%(v/v) glycerol |
| |
|
|
| Optimized crystal mounted in a cryo-loop |
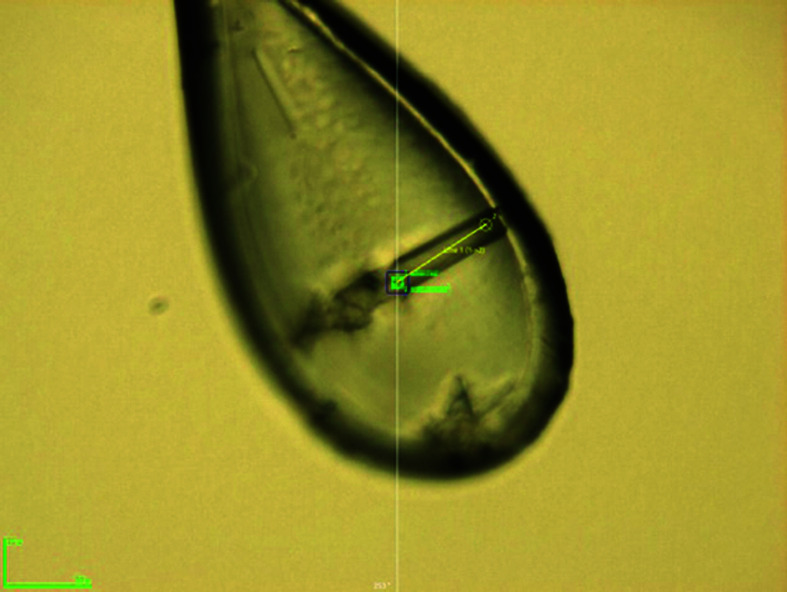
|
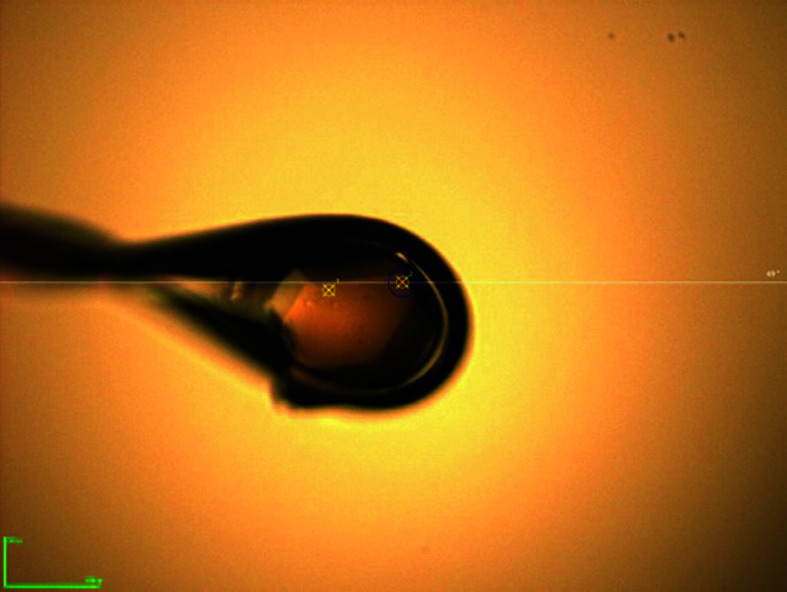
|
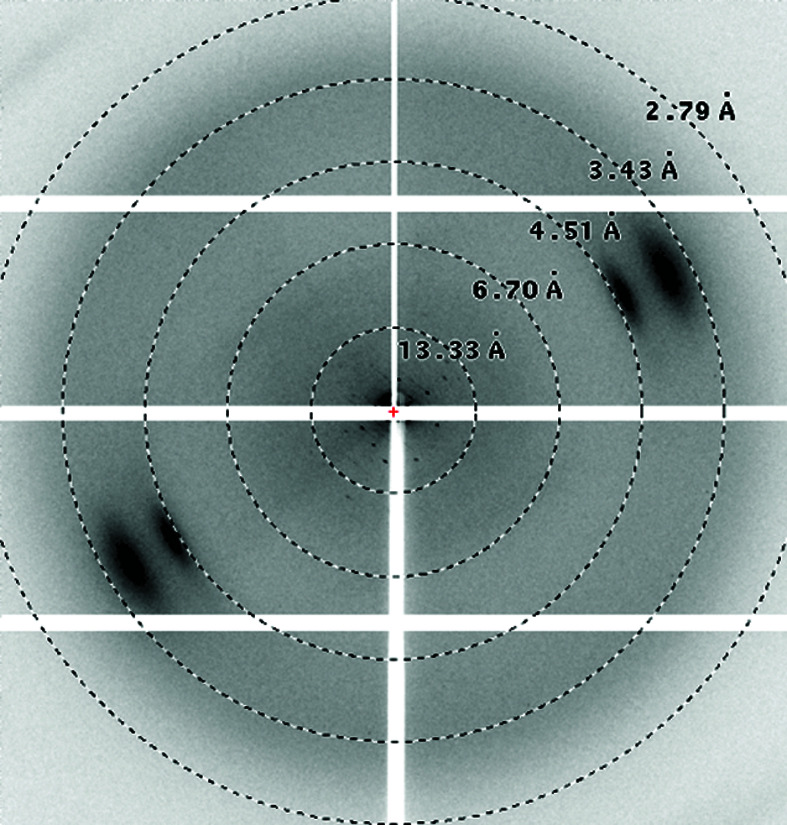
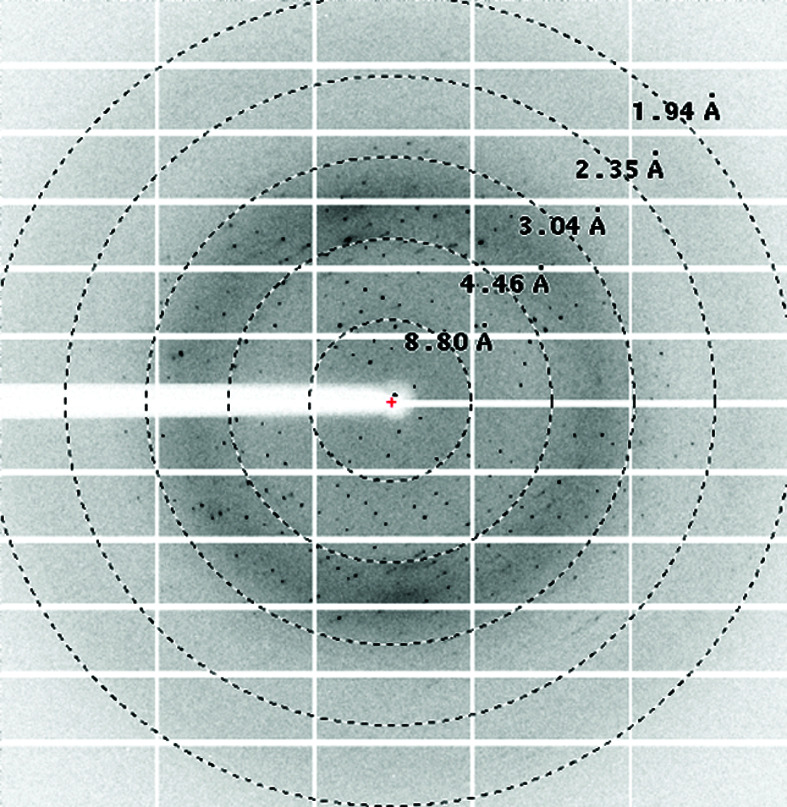
| Diffraction pattern |
|
|
| Diffraction limit (Å) |
∼8 |
1.85 |






