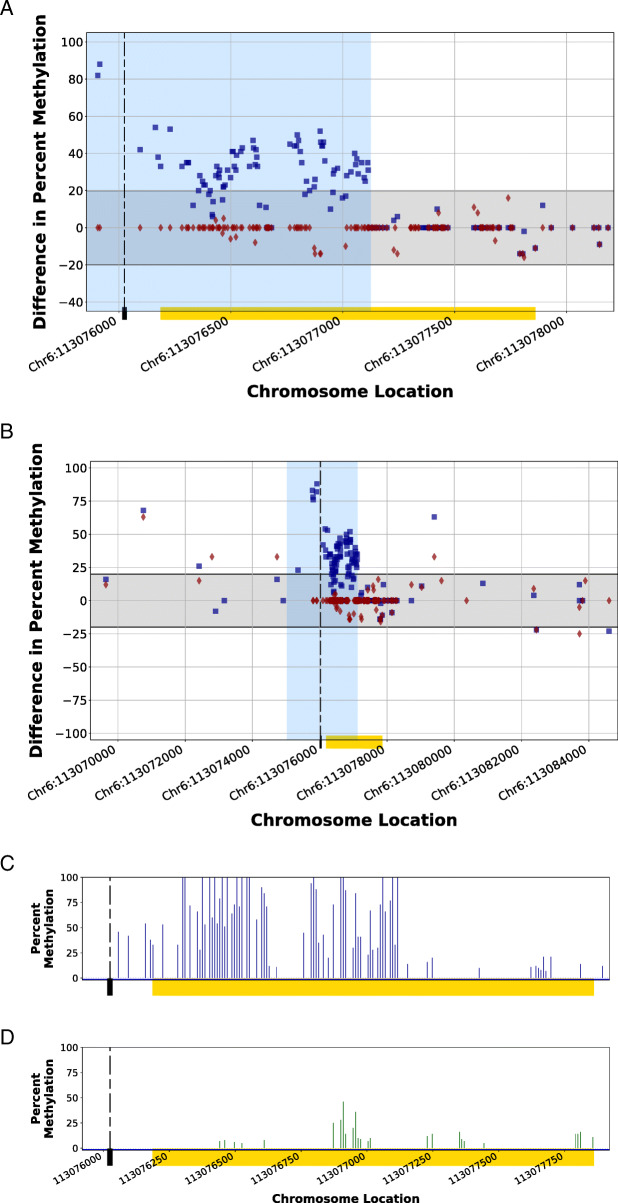
Fig. 6.
Methylomic difference of a partially edited CpG island within the genome of the HDR2 Animal using homology-directed repair in a CRISPR-mediated edit. The downstream homology arm of a synthetic DNA fragment (region shaded in blue) in a homology-directed repair of a CRISPR-mediated genomic cut spanned a portion of a CpG island (CGI; Chr6:113,076,186–113,077,861; yellow bar) within the genome of the HDR2 Animal, resulting in a destabilized methylation pattern (blue squares) as compared to the Control 1 Animal methylation patterns at the same location (red diamonds). The regions with methylation patterns modified by the incorporation of homology arms within the CGI and upstream of the CGI are shaded in a light blue background. While Fig. 5a illustrates the localized methylation variance introduced at the CGI, Fig. 5b illustrates the comparison of the modified region to flanking endogenous genomic regions by displaying the variance of methylation patterns for 7000 bp upstream and downstream of the CGI. The methylation patterns of the unmodified downstream portion of the CGI (unshaded) for the edited CGI as well as the flanking regions of endogenous genome displayed a methylation pattern similar to the control. The gray band indicates considered biological epigenetic variance (+/− 20% change). The dashed line indicates the protospacer adjacent motif (PAM) location of the targeted CRISPR cut site. The comparison of the percent methylation of the CpG sites observed at the localized CGI region for the CRISPR-edited animal (Fig. 5c) and the unedited control animal (Fig. 5d) demonstrates the introduced methylation variance as a result of the adoption of the donor DNA. Blue squares (■) indicate the percent differences in CpG methylation for HDR2 Animal from Control 2 Animal at given chromosome locations. Red diamonds (♦) indicate the percent differences in CpG methylation for Control 1 Animal from Control 2 Animal at given chromosome locations