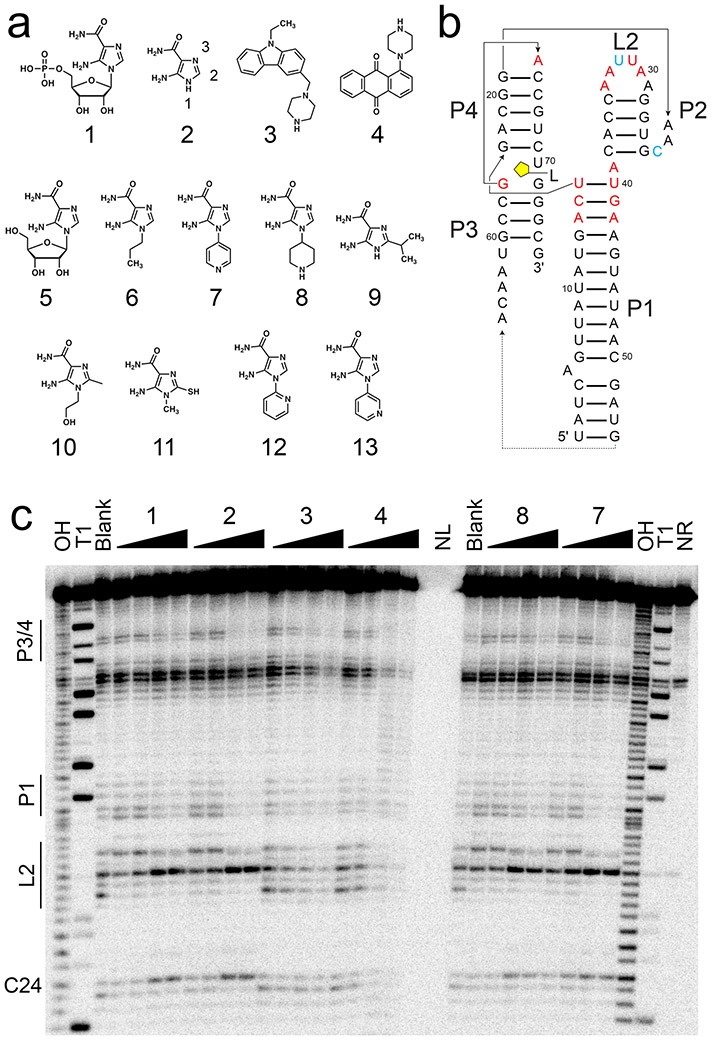
Figure 1.
In-line probing of ZTP riboswitch with AICA derivatives and SMM hits. (a) Chemical structures of compounds used herein—1, 5-aminoimidazole-4-carboxamide-β-1-D-ribofuranoside 5’-monophosphate (ZMP); 2, 5-aminoimidazole-4-carboxamide (AICA), with functional group numbering as indicated; 3, 9-ethyl-3-(1-piperazinylmethyl)-9H-carbazole; 4, 1-(1-piperazinyl)anthra-9,10-quinone; 5, AICA-β-1-D-ribofuranoside (AICAR); 6, 1-propyl-AICA; 7, p-1-pyridinyl-AICA; 8, p-1-piperidinyl-AICA; 9, 2-isopropyl-AICA; 10, 1-hydroxyethyl-2-methyl-AICA; 11, 1-methyl-2-mercapto-AICA; 12, o-1-pyridinyl-AICA; 13, m-1-pyridinyl-AICA. (b) Secondary structure of the F. ulcerans ZTP riboswitch used for in-line probing is shown, with the approximate ligand binding site indicated in yellow. Paired helical elements P1-4 and loop L2 are noted. Nucleotides with decreased cleavage (red) and increased cleavage (cyan) in the presence of 1, 2, 7, and 8 are indicated. (c) On the left side of the gel, regions of the ZTP riboswitch most sensitive to changes in in-line probing reactivity are noted. From left to right, the lanes are: OH, partial alkaline digest; T1, RNase T1 digest; Blank, no-compound control; 1 at 3, 6, 200, 400 nM; 2 at 1.5, 3, 100, 200 μM; 3 at 16, 31, 1000, 2000 μM; 4 at 16, 31, 1000, 2000 μM; NL, no loading; 8 at 0.16, 0.31, 10, 20 μM; 7 at 0.16, 0.31, 10, 20 μM; and NR, no reaction. Example gel is shown (n = 6 independent experiments). See also Figures S1 and S2.