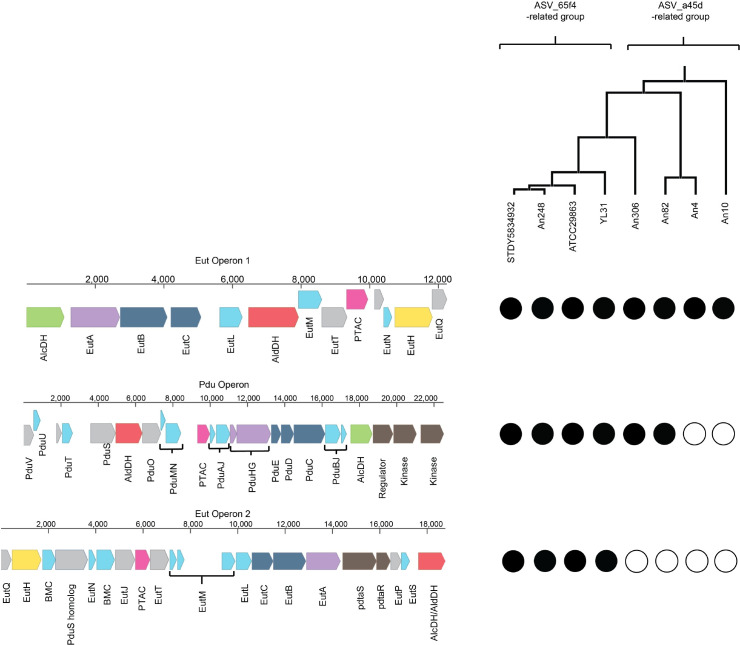
Fig 6. Representative BMC Loci for Flavonifractor spp..
The ethanolamine utilization operons (Eut operon 1 and 2) and 1,2-propanediol utilization operon (Pdu operon). Genes are drawn on the F. plautii YL31 genome using Benchling [Biology Software] (2019). Eut operon 1 is 12,247 bp, Pdu operon is 22,527 bp, and Eut operon 2 is 18,769 bp. Abbreviations are as follows: AlcDH, Alcohol dehydrogenase; AldDH, Aldehyde dehydrogenase; PTAC, phosphotransacetylase; BMC, bacterial microcompartment; PdtaS, two-component system, sensor histidine kinase; PdtaR, two-component system, response regulator. Genes are color-coded according to their annotation: light blue, BMC-containing proteins; red, aldehyde dehydrogenase; green, alcohol dehydrogenase; solid pink, PduL-type phosphotransacylase; light purple, re-activating proteins; dark blue, signature enzymes (ethanolamine ammonia lyase subunits and propanediol dehydratase subunits); brown, regulatory element including two-component signaling elements; yellow, transporter; gray, other Eut or Pdu proteins. Circles show the presence (filled circle) or absence (white circle) of proteins in the strains depicted in the phylogenetic tree. The UPGMA phylogenetic tree involved 8 nucleotide sequences: F. plautii 2789STDY5834932 (STDY5834932), F. plautii An248 (An248), F. plautii ATCC 29863 (ATCC29863), F. plautii YL31 (YL31), Flavonifractor sp. An306 (An306), Flavonifractor sp. An82 (An82), Flavonifractor sp. An4 (An4), and Flavonifractor sp. An10 (An10). All positions containing gaps and missing data were eliminated. There were a total of 1008 positions in the final dataset. S8 Table contains all accession numbers for each protein in each strain. Pdu operon was reconstructed by its similarity with the one in Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (S9 Table).