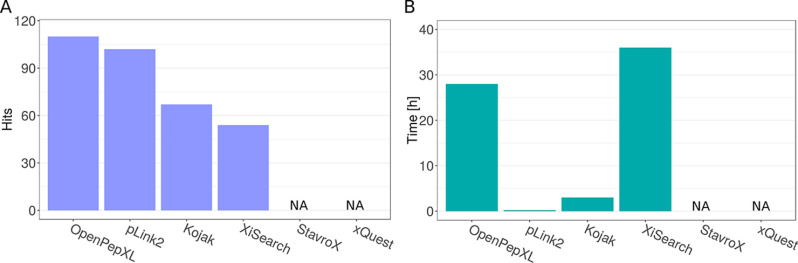
Fig. 3.
Results from the analysis of the ribosomal fraction data set. A, Numbers of identified unique residue pairs (URPs) in the ribosomal fraction data set with a target database of 128 proteins. OpenPepXL identified 110 URPs, pLink2 identified 102 URPs, Kojak 67 URPs and XiSearch 54 URPs. Structural verification of these cross-links is presented in supplemental Figs. S3 and S4. StavroX exceeded the available memory of 8 GB and could not finish the search. xQuest did not exceed the available memory, but the search was canceled because the projected runtime under these conditions was unreasonable. B, Runtimes in hours needed to analyze the ribosomal fraction data set with a database of 128 target and 128 decoy proteins using one CPU core. pLink2 only took 15 min. Kojak took 3 h, OpenPepXL 28 h and XiSearch 36 h.