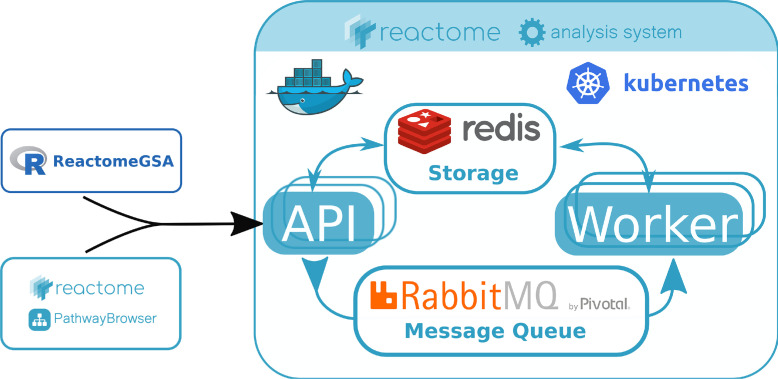
Fig. 1.
Schema of the ReactomeGSA system. All requests are sent to a public web-based API through the ReactomeGSA Bioconductor R package or Reactome's web-based PathwayBrowser. The system is a Kubernetes application based on the microservices architecture. All requests are distributed through an internal message queue using RabbitMQ. Worker nodes are responsible for the complete pathway analysis, including identifier mapping and the creation of the visualization data in Reactome's pathway browser. Data nodes are responsible to load data from external resources such as ExpressionAtlas. Finally, report nodes create PDF and Microsoft Excel files as a static report of the analysis results. All data are stored in a central Redis instance. All nodes are Docker containers that are orchestrated by Kubernetes and automatically scaled based on current demand. Thereby, the application can dynamically adapt to changing usage levels.