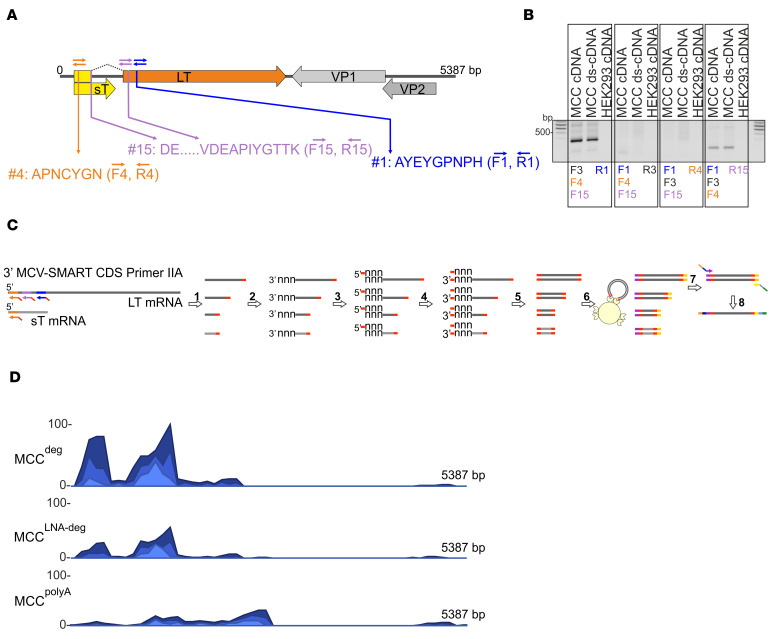
Figure 2. dMS-identified peptides facilitate identification of viral sequences by NGS with cDNA libraries generated using degenerate oligonucleotides.
(A) Schematic illustration of the MCV genome. Early (LT, yellow and orange; sT, yellow) and late (VP1, light gray; VP2, dark gray) region open reading frames are shown. The corresponding positions of the 3 MCV peptides identified by dMS (features 4, 15, and 1) and degenerate primer binding sites are shown in orange, purple, and blue arrows, respectively. (B) RNA extracted from MCC tissue (tissue R16–67) or HEK293 cells were subjected to cDNA synthesis with random hexamers and, additionally, second-strand synthesis for the MCC sample (double-stranded cDNA; ds-cDNA). cDNAs were amplified using the indicated combinations of degenerate primers (Supplemental Table 3) corresponding to the peptide sites highlighted in light blue (F1, R1), violet (F15, R15) and orange (F4, R4). F, forward; R, reverse. F3 and R3 (black) are non-MCV primers. (C) Library generation using SMART oligonucleotides and Nextera DNA Flex. Step 1: 3′ SMART CDS Primer IIA (Supplemental Table 4) mediated first-strand synthesis. Step 2: Tailing by RT. In the cDNA reaction, nontemplated bases (nnn) are added to the ends of nascent cDNA by the terminal transferase activity of RT. Step 3: SMARTer IIA oligo anneals to nontemplated bases at cDNA ends (nnn). Step 4: Template switch and extension at 3′ end. The RT polymerase switches strands to transcribe the complement of the oligonucleotide, leaving the SMART adaptor at both ends of cDNA. Step 5: Long-distance PCR with single 5′ PCR Primer IIA amplifies libraries. Step 6: Bead-linked transposomes mediate the simultaneous fragmentation of ds-cDNA and the addition of Illumina sequencing primers using Nextera DNA Flex. Step 7: Reduced-cycle PCR amplification amplifies sequencing-ready DNA fragments and adds indexes and adapters. Step 8: Sequencing-ready fragments are washed and pooled. (D) NGS coverage maps of MCC RNA-seq libraries. RNA-seq reads were obtained from 3 different samples to compare the efficiency of MCV read recovery using various primer pool sets for cDNA and library generation (Supplemental Table 3). Ribo-depteted MCC RNA (R11–65) was subjected to cDNA synthesis with SMART-degenerate oligo pool (MCCdeg), LNA modified SMART-degenerate oligo pool-SMART (MCCLNA-deg), and modified oligo-dT-SMART (MCCpolyA) and then subjected to library generation using Nextera DNA Flex application. Standardized coverage depths (reads) for comparison purposes are indicated on the y axis for each alignment.