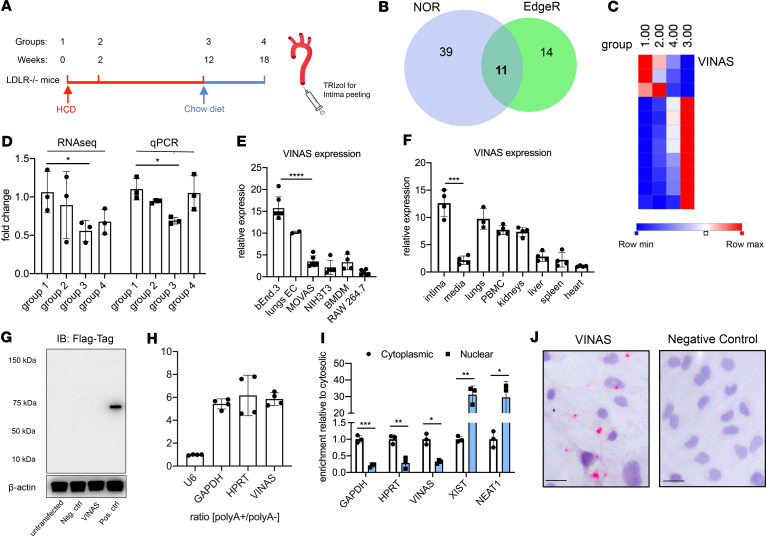
Figure 1. Identification of the lncRNA VINAS in lesional intima.
(A) RNA derived from aortic intima of LDLR–/– mice (n = 3; each sample represents RNA pooled from 2 mice) that were placed on a high-cholesterol diet (HCD) for 0 (group 1), 2 (group 2), 12 (group 3), and 18 weeks after 6 weeks of resumption of a normal chow diet (group 4). (B) Venn diagram displays significantly dysregulated lncRNAs in genome-wide RNA-Seq profiling using EdgeR and no-overlapping reads (NOR) showing intersecting hits (n = 11), uniquely identified in EdgeR (n = 14) or NOR (n = 39), (log2 fold change [1.5]; FDR < 0.05). (C) Heatmap for 11 lncRNAs that were dynamically regulated with progression and regression of atherosclerosis (n = 3). (D) RNA-Seq results for VINAS across groups 1–4 obtained by RNA-Seq analysis and verified by RT-qPCR (n = 3). (E) RT-qPCR expression analysis for VINAS in different cell types (n = 3). (F) VINAS expression in body organs and PBMCs of 24-week-old C57BL/6 mice (n = 4). (G) To test the coding potential, VINAS sequence was cloned upstream of 3xFlag-Tag cassette, transfected in HEK293T cells, and immunoblotted for Flag antibody. Positive control was provided with the kit (representative of 3 experiments). (H) RNA from mouse extracellular cells (ECs) was isolated for polyA+ and polyA– enriched RNA and analyzed by real-time quantitative PCR (RT-qPCR) (n = 3). (I) RT-qPCR analysis for RNA derived from mouse ECs separated into cytoplasmic and nuclear fractions and normalized to the cytoplasmic fraction (n = 3). (J) RNA in situ hybridization for negative control and VINAS probes on paraformaldehyde-fixed mouse ECs. Scale bar: 5 μm. Data represent the mean ± SD. Statistical differences were calculated using unpaired 2-tailed Student’s t test except for multiple comparisons (E and F) in which 1-way ANOVA with Bonferroni’s correction was used. *P < 0.05, **P < 0.01, ***P < 0.001.