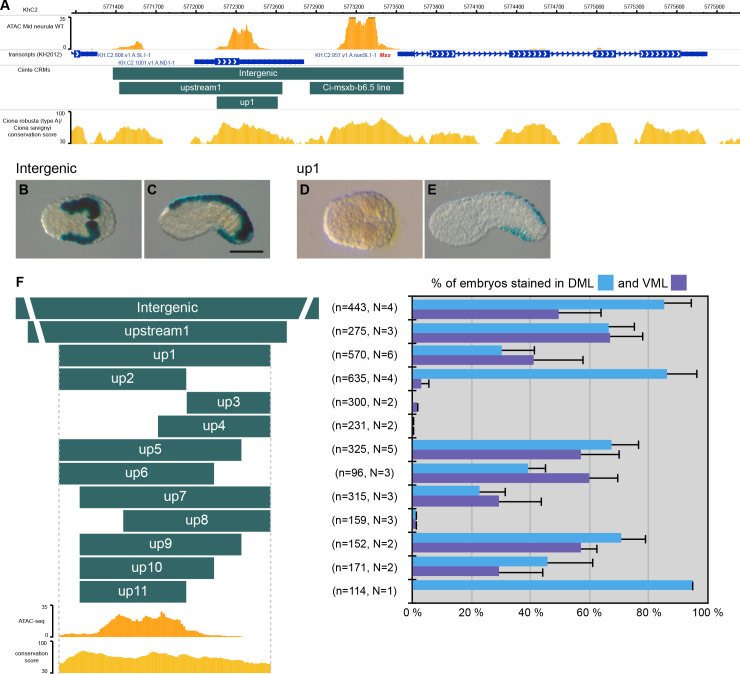
Figure 1. CRMs controlling Ciinte.Msx expression in VDML.
(A) Snapshot of the Ciinte.Msx locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between C. robusta and C. savignyi (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). (B–E) Representative examples of X-gal stained embryos at late gastrula stages (B, D) and early tailbud stages (C, E) following C. intestinalis embryos electroporation of Ciinte.Msx-Intergenic (B, C) and Ciinte.Msx-up1 (D, E). Embryos are shown in dorsal view (B, D) and in lateral view with dorsal to the top (C, E), and anterior to the left. Scale bar: 100 μm. (F) Schematic representation of the various constructs and their activity at early tailbud stages in DML (blue) and VML (purple) (n indicates the total number of embryos examined; N indicates the number of independent experiments).