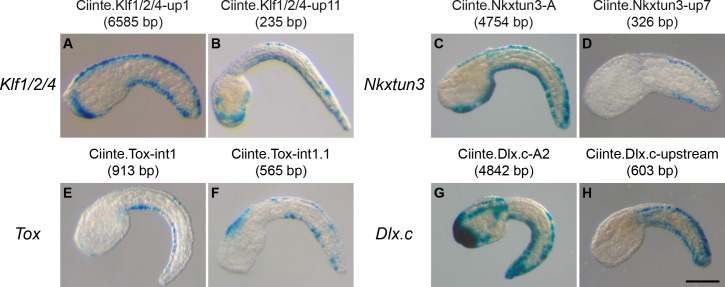
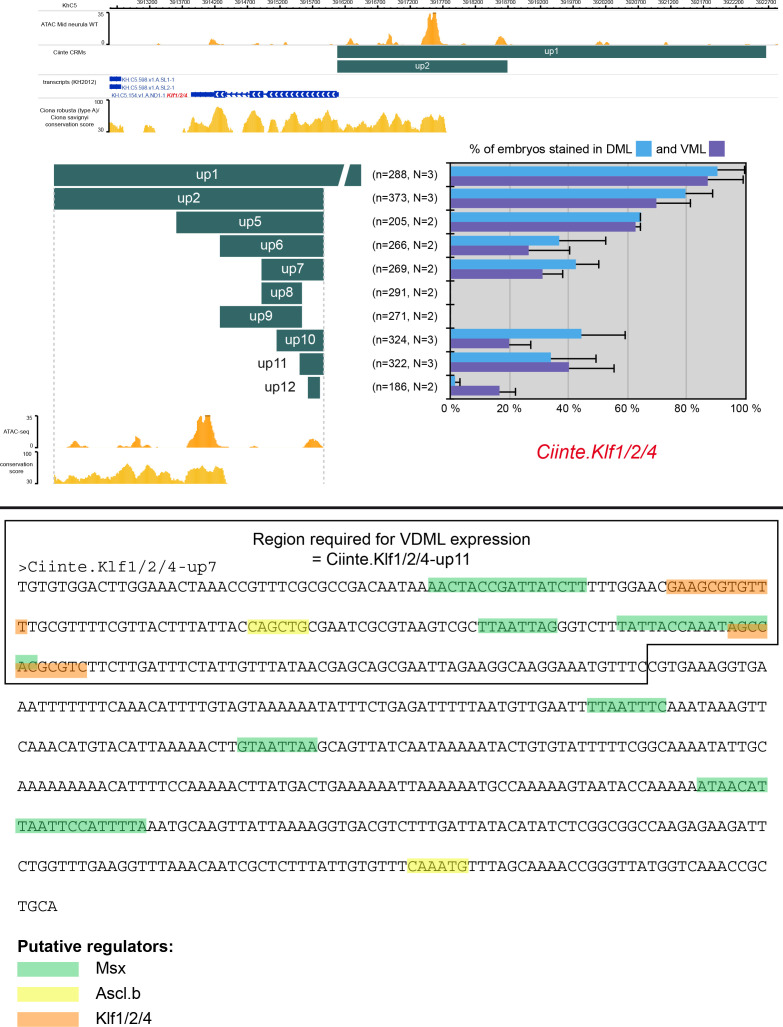
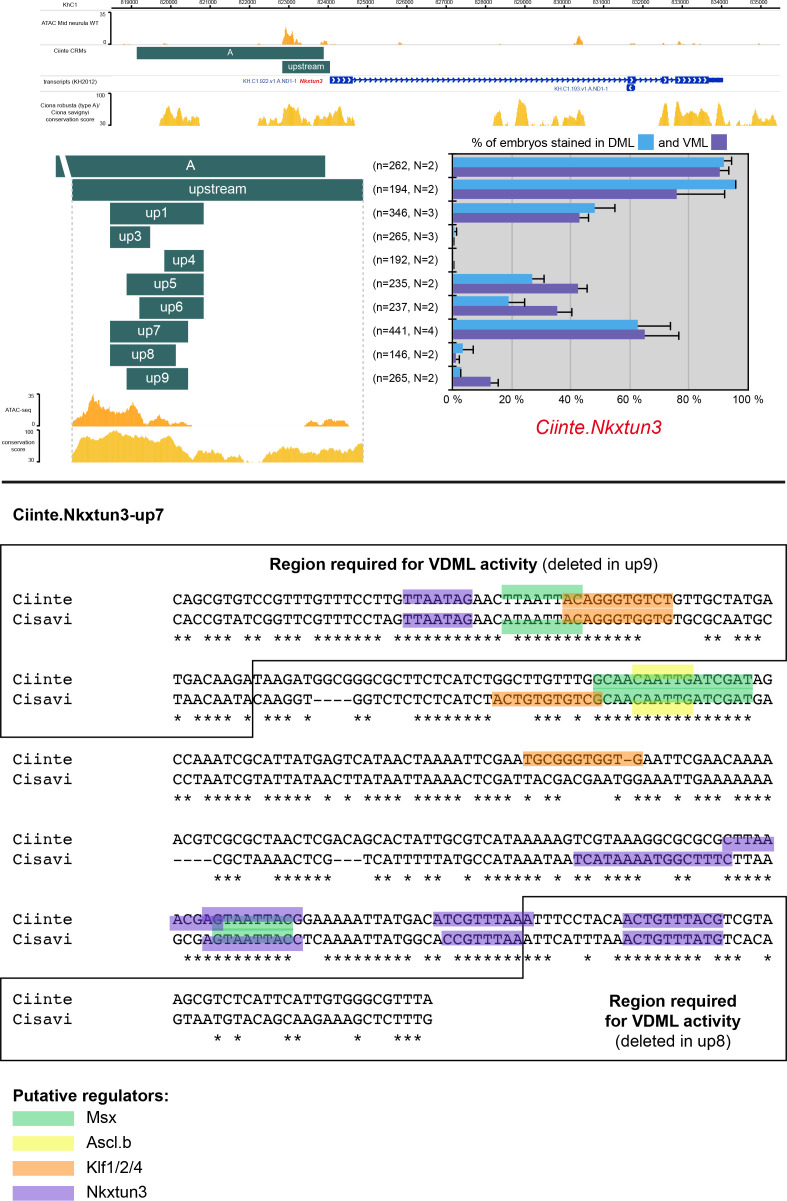
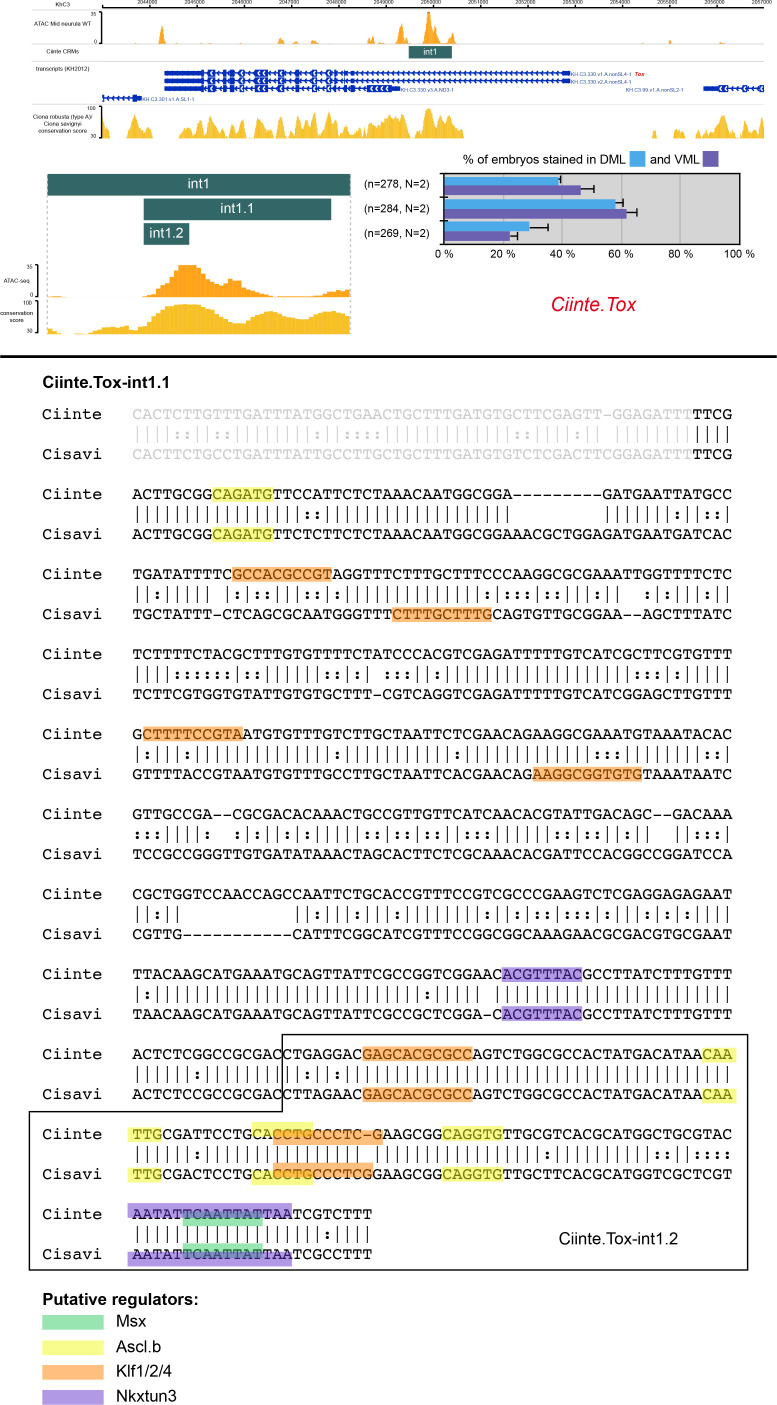
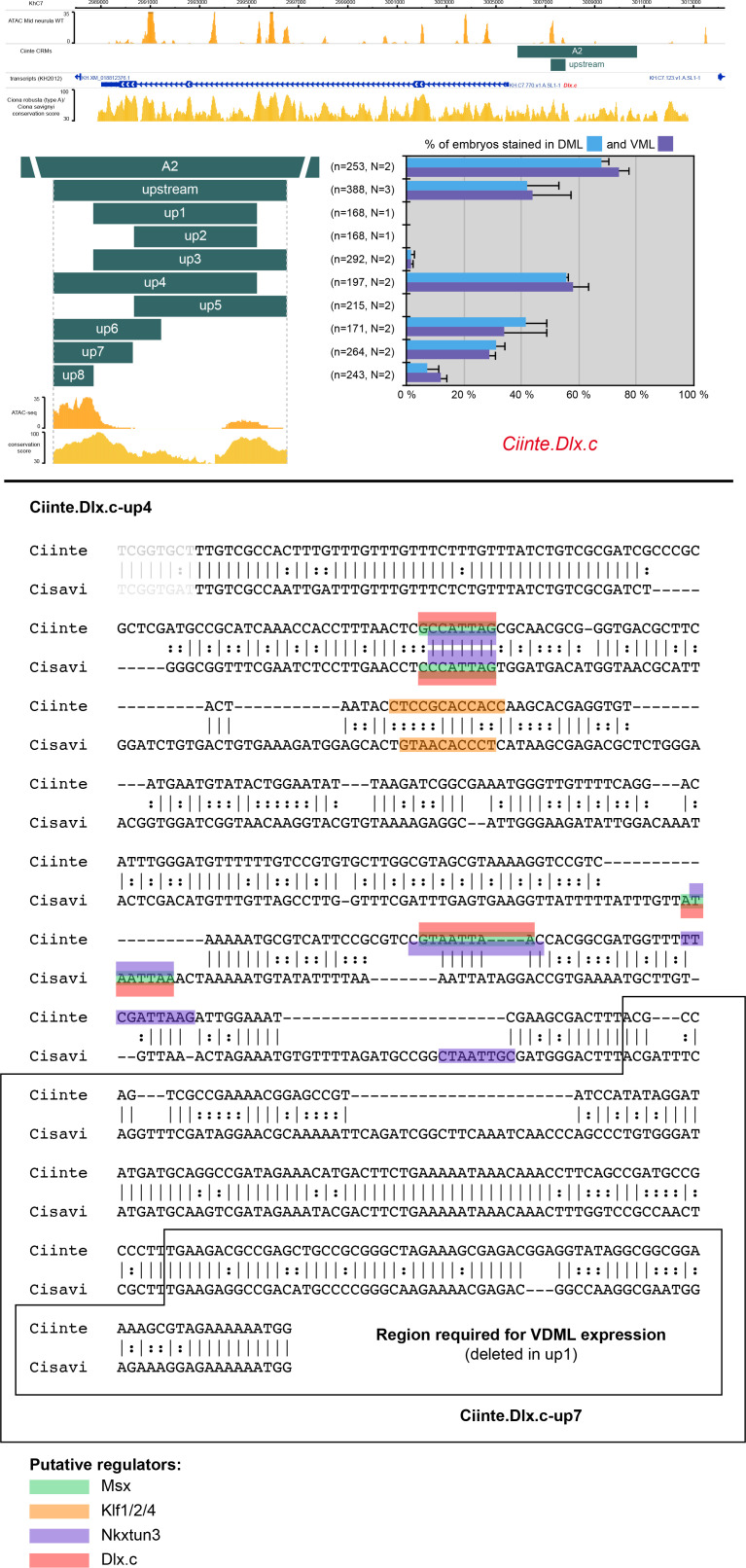
Figure 3. CRMs with activity in the tail epidermis midlines for Ciinte.Klf1/2/4, Ciinte.Nkxtun3, Ciinte.Tox, and Ciinte.Dlx.c.
Representatives examples of X-gal stained embryos at tailbud stages following C. intestinalis embryos electroporation of C. intestinalis genomic regions for Klf1/2/4 (A, B), Nkxtun3 (C, D), Tox (E, F) and Dlx.c (G, H). For each gene, an example for the largest and the smallest regions with robust VDML activity are shown (the size of the region is shown between parentheses after the CRM's name). Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 100 μm.