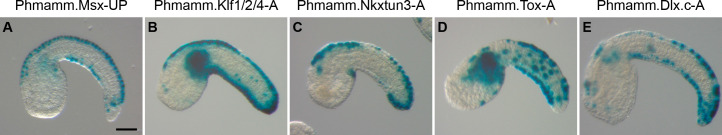
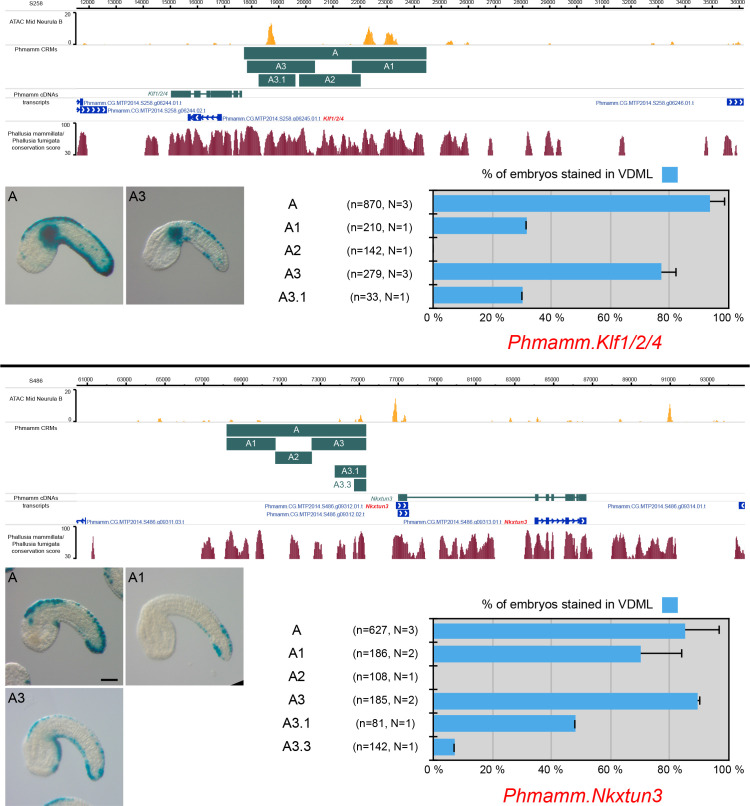
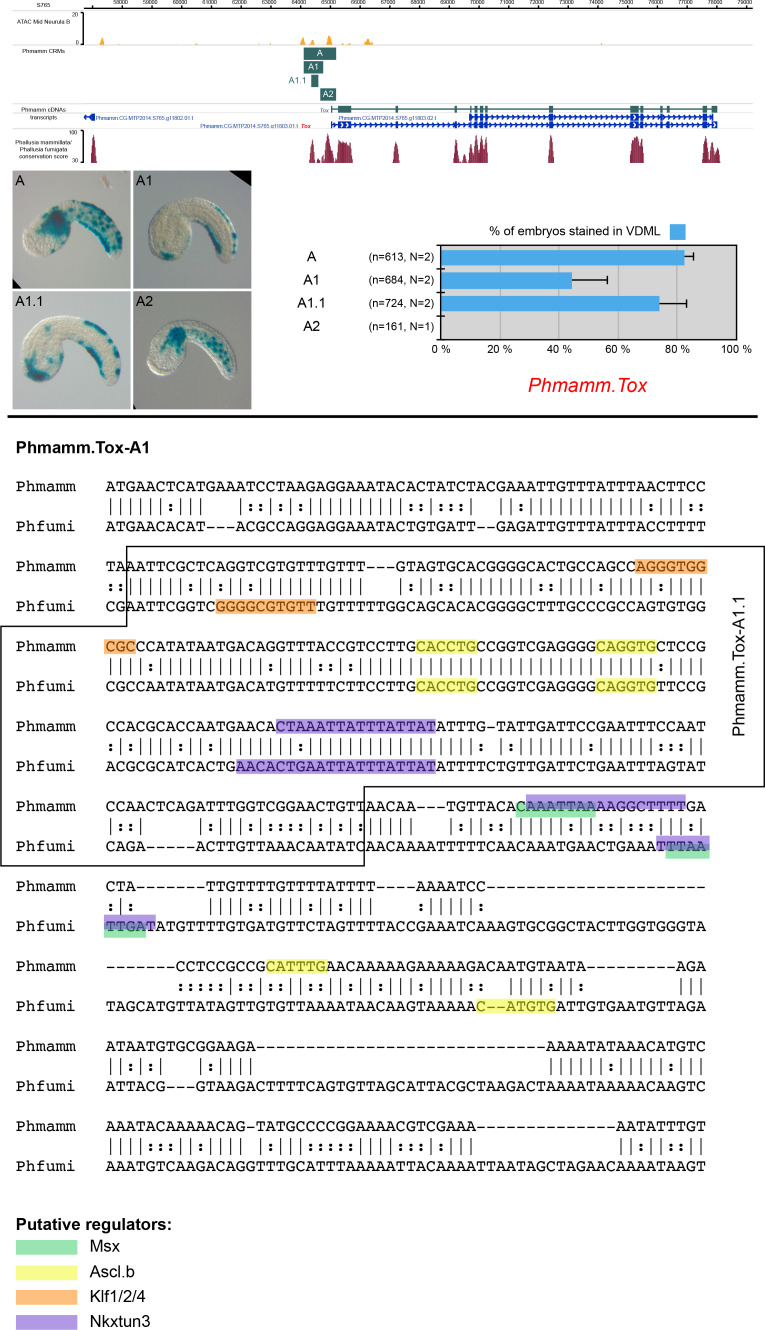
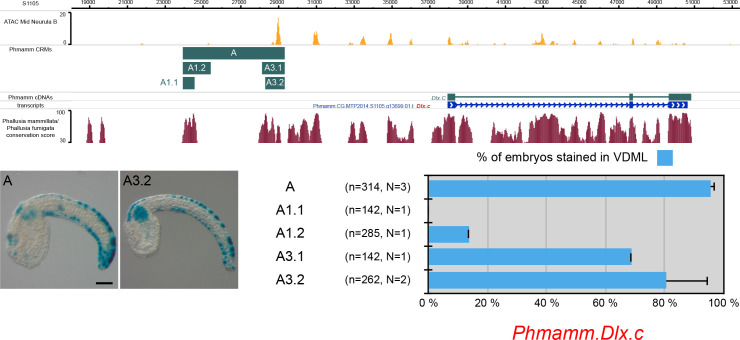
(Top) Snapshot of the
Phmamm.Msx locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between
P. mammillata and
P. fumigata (from
https://www.aniseed.cnrs.fr/ and
Dardaillon et al., 2020;
Madgwick et al., 2019). Representative examples of X-gal staining at tailbud stages following
P. mammillata embryos electroporation of
P. mammillata genomic regions for
Msx (Phmamm.Msx-UP (same picture as
Figure 5A) and Phmamm.Msx-LED). Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 50 μm. Schematic representation of the various constructs and their activity at tailbud stages in VDML (blue) (n indicates the total number of embryos examined, N indicates the number of independent experiments). (Bottom) Identification of putative TFBS for candidate upstream factors in Phmamm.Msx-LED aligned with its counterpart from
P. fumigata. All putative sites for ventral factors (SBE, BRE, Tbx2/3, Nkxtun1, Nkx2-3/5/6 and Irx.c) and dorsal factors (Msx and Su(H)/Rbpj) have been mapped, but only conserved sites are shown: 5 SBE, 2 BRE, 1 Nkxtun1, 1 Nkx2-3/5/6, and 1 Msx sites. When comparing with Ciinte.Msx-up10 (
Figure 1—figure supplement 2), we found shared sites for direct activation by Bmp signaling (SBE and BRE), and for activation by Msx itself. However, sites for ventral TFs were different: Nkxtun1 and Nkx2-3/5/6 in Phmamm.Msx-LED and Irx.c and Tbx2/3 in Ciinte.Msx-up10. Note that the size of the highlighted site for of a given TF may vary depending on the matrix used.