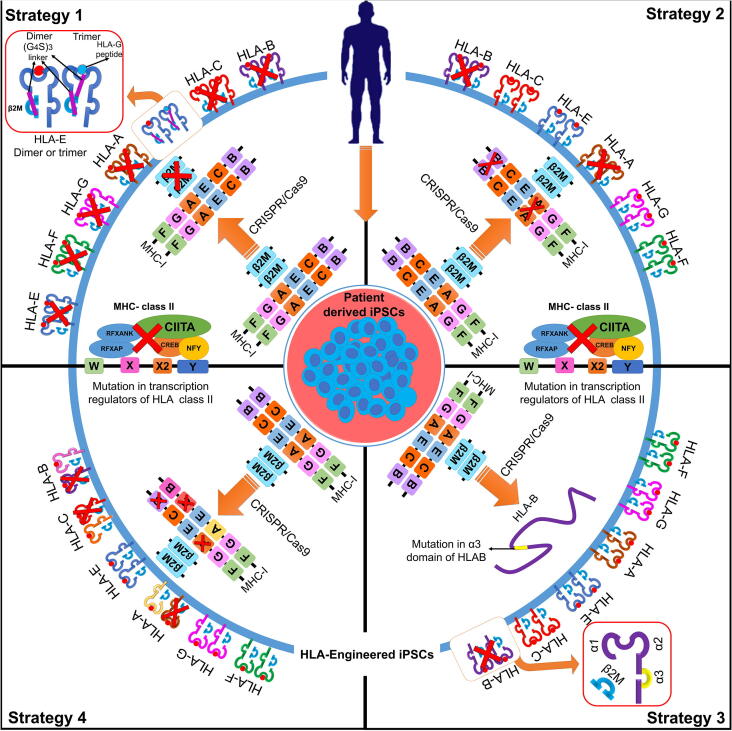
Fig. 2.
CRISPR-based HLA immunoengineering toward development of immunocompatible-induced pluripotent stem cells. The figure represents various strategies aimed towards generation of HLA-immunocompatible iPS cell lines by selectively targeting HLA class I and/or II using the CRISPR/Cas9 system. Strategy 1: Selective knockout of transactivators for HLA class I-B2M gene and HLA class II-CIITA using a CRISPR/Cas9 system prevents immune response from cytotoxic CD8+ and CD4+ T cells [229]. However, lack of HLA surface expression can activate NK cells, risking proliferation of infected cells and increasing the risk of cancer. Exogenous surface expression of HLA E single-chain dimers fused to B2M promoter or as trimers containing HLA G signal peptides in cells lacking surface expression of HLA-A, B or C escapes allogenic recognition by CD8+ T cells while conferring resistance to NK cell mediated lysis [236]. Strategy 2: HLA-C plays a pivotal role in suppressing activation of NK cells. CRISPR/Cas9-mediated disruption of HLA-A and HLA-B biallelically, while retaining HLA-C along with other non-canonical HLA class I in iPSCs expands donor compatibility while suppressing T cell and NK cell activation. The immunocompatibility of HLA-C-retained iPSCs can be improved by targeted knockout of class II transactivator-CIITA, suppressing activation of CD4+ T cells [234]. Strategy 3: The CRISPR/Cas9 system has been demonstrated to increase immunocompatibility by knocking out heterozygous HLA-B in homozygous HLA-A iPSCs with an HLA haplotype HLA-A (24:02, 24:02) and HLA-B (40:01, 54:01). HLA-B-engineered iPSCs were generated by specifically mutating the alpha 2–3 domains that form the B2M binding groove. The HLA-KO homozygous iPSCs exhibit less immunogenicity while maintaining pluripotency [237]. Strategy 4: Generation of pseudo-homozygous iPSCs using the CRISPR/Cas9 system. HLA homozygous cell line stocks have been proposed to match a major percentage of the population [226]. CRISPR/Cas9 gene editing can be used to generate class I HLA haploid iPSCs by targeted allele-specific HLA disruption in HLA heterozygous iPSCs to generate “pseudo-homozygous” iPSCs with similar donor potential to HLA homozygous iPSCs [234].