Abstract
Immune checkpoint blockade (ICB) has shown remarkable clinical efficacy in several cancer types. However, only a fraction of patients will respond to ICB. Here, we performed pooled mutagenic screening with CRISPR-mediated genetically engineered mouse models (CRISPR-GEMMs) in ICB settings, and identified KMT2D as a major modulator of ICB response across multiple cancer types. KMT2D encodes a histone H3K4 methyltransferase and is among the most frequently mutated genes in cancer patients. Kmt2d loss led to increased DNA damage and mutation burden, chromatin remodeling, intron retention, and activation of transposable elements. Additionally, Kmt2d-mutant cells exhibit increased protein turnover and IFN-γ-stimulated antigen presentation. In turn, Kmt2d-mutant tumors in both mouse and human are characterized by increased immune infiltration. These data demonstrate that Kmt2d deficiency sensitizes tumors to ICB by augmenting tumor immunogenicity, and also highlight the power of CRISPR-GEMMs for interrogating complex molecular landscapes in immunotherapeutic contexts that preserve the native tumor microenvironment.
Introduction
Checkpoint immunotherapy has achieved substantial success, showing clinical benefits across multiple tumor types with durable responses even in chemo-resistant and metastatic cancers (1–4). However, the majority of patients do not respond to checkpoint immunotherapy (5,6), indicating the importance of precision immunotherapy – where patients are stratified based on functional and clinical evidence, subsequently receiving the treatments or combinations most likely to benefit them.
A multitude of approaches have been applied to understand the features associated with immunotherapy response (7,8). These include whole-genome sequencing (7,9,10), proteomics analysis (11), single-cell transcriptomic analysis (12), in vitro cancer-immune cell co-cultures (13,14), and ex vivo / in vivo screens using cell lines in tumor transplant models (15). Several factors, including PD-L1 expression, tumor mutation burden (16), neoantigen burden (17), immune infiltration status (18,19), as well as certain oncogenic pathways (20) have been demonstrated to be correlated with immunotherapy response. Additionally, many mechanisms have been described in primary or acquired resistance to immunotherapy (21,22). For instance, tumors can foster the development of an immunosuppressive tumor microenvironment (23), or acquire new mutations that reduce immune recognition and apoptosis (24).
Despite these advances, our understanding of the genetic factors that dictate response to checkpoint immunotherapy remains incomplete. Analysis of patient cohorts can reveal associations with ICB response, but such studies cannot firmly establish causality. Current genetic screening approaches using in vitro or ex vivo cultured cell lines are confined by the mutation background, and may miss subtle factors that influence ICB response in the complex immunological setting of the tumor microenvironment. Genetically engineered mouse models (GEMMs) (25) can more precisely mimic the features of human cancers, because such tumors develop from cells within the native organs of fully immunocompetent animals, thereby preserving the immune microenvironment. Because of these features, GEMMs offer certain distinct advantages for the studies of tumor immunology. While traditional GEMMs can only target a handful of genes at a time, CRISPR enables pooled targeting of multiple genes through somatic genome editing. We have previously developed CRISPR-GEMMs that enabled large-scale direct in vivo screening of functional tumor suppressors (26,27). Using CRISPR-GEMMs, genetically complex tumors can be readily generated in individual mice that each reflect the genetic and cellular heterogeneity of human tumors, with the flexibility to target any desired sets of genes.
Here, we performed a CRISPR-GEMM screen of significantly mutated genes (SMGs) in human cancers (28,29), examining the effect of these mutations on ICB response. We specifically pinpoint Kmt2d deficiency as a major mediator of sensitivity to ICB therapy in diverse cancer types, suggesting its potential as a biomarker for patient stratification.
Results
A CRISPR-GEMM screen identifies genetic modulators of immunotherapy response in vivo
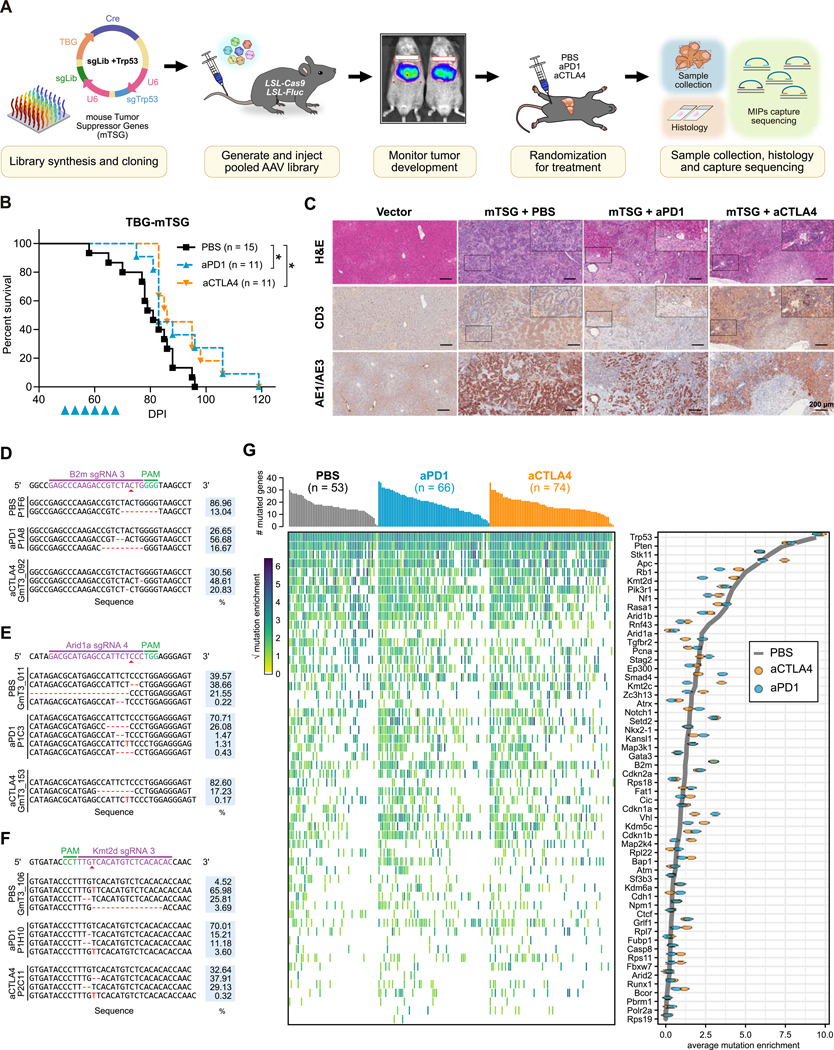
To perform a screen for genetic modulators of immunotherapy response in conditions that closely mimic human cancers, we developed a CRISPR-GEMM model of liver cancer, in which AAV-CRISPR-mediated pooled mutagenesis drives autochthonous liver tumorigenesis in fully immunocompetent mice. We designed an AAV-CRISPR vector that expresses Cre recombinase under a liver-specific thyroxine binding globulin (TBG) promoter, together with two sgRNA expression cassettes: one for a Trp53-targeting sgRNA, and the other as a backbone sgRNA designed for cloning and expression of additional specific sgRNA(s). We utilized the mTSG library, which targets the top 49 most frequently mutated tumor suppressor genes in the pan-cancer TCGA datasets with 7 housekeeping genes as internal controls (26,27). We cloned the mTSG library into the AAV-TBG-CRISPR vector, and generated AAVs carrying the pooled sgRNA library (Fig. 1A). In order to monitor liver tumorigenesis in vivo, we crossed LSL-Cas9–2A-EGFP (LSL-Cas9) mice with LSL-firefly luciferase (LSL-Fluc) mice to generate LSL-Cas9; LSL-Fluc mice. We then introduced the base vector AAVs (AAV-Vector), sgTrp53-delivering AAVs (AAV-sgTrp53) or mTSG library AAVs (AAV-mTSG) into the mice by intravenous injection.
Figure 1. AAV-CRISPR direct in vivo screening to pinpoint genetic modulators of immunotherapy response.
(A) Schematic of the experimental design. An sgRNA library targeting the murine homologs of the 49 most frequently mutated tumor suppressor genes, along with 7 housekeeping genes (mTSG; 288 sgRNAs) was cloned into an AAV-CRISPR vector containing a liver-specific Cre expression cassette and a Trp53-targeting sgRNA. AAVs were produced and injected intravenously into LSL-Cas9; LSL-Fluc mice. The mice were assigned into 3 groups based on luciferase imaging, then received anti-PD1 (aPD1), anti-CTLA4 (aCTLA4), or PBS treatment. Tumors were processed for histology and MIP capture sequencing to profile the mutational landscape of all targeted genes.
(B) Kaplan-Meier survival curves of AAV-mTSG injected mice, treated with PBS (black, n = 15), aPD1 (blue, n = 11), or aCTLA4 (orange, n = 11) mice. All PBS treated mice died within 3 months, while aPD1 treated mice (P = 0.0389) and aCTLA4 treated mice (P = 0.0185) had longer survival (log-rank test).
(C) Representative images of hematoxylin and eosin (H&E), CD3, and AE1/AE3 staining of liver sections from AAV-Vector or AAV-mTSG injected mice, treated with PBS, aPD1, or aCTLA4. Scale bar is 200 μm.
(D-F) Representative insertions and deletions (indels) observed at the genomic region targeted by B2m sgRNA3 (D), Arid1a sgRNA4 (E), and Kmt2d sgRNA 3 (F) in mTSG-treated samples from PBS, aPD1, or aCTLA4 treatment groups. The percentage of each variant is indicated on the right.
(G) Mutational landscape of AAV-mTSG liver tumors (PBS, n = 53; aPD1, n = 66; aCTLA4, n = 74). Top, bar plot of the number of mutated genes in each sample. Center, heat map of mutation enrichment scores for each of the 56 targeted genes across all samples. Right, dot plot of the average mutation enrichment score for each gene, grouped by treatment condition (PBS, gray line; aPD1, blue; aCTLA4, orange).
Asterisks: * P < 0.05, ** P < 0.01, *** P < 0.001.
See also: Supplementary Fig. S1
We monitored the bioluminescence signals in the injected mice using the intravital imaging system (IVIS). We observed a steady increase of luciferase signal from day 40 to day 60, indicative of ongoing tumorigenesis (Supplementary Fig. S1A). Based on the IVIS data, we assigned the AAV-mTSG injected mice into 3 size-matched cohorts to receive PBS, anti-PD1, or anti-CTLA4 treatment (Fig. 1A). All of the AAV-mTSG injected mice treated with PBS died within 100 days (Fig. 1B), having developed large liver tumors with 100% penetrance. In sharp contrast, no mice died from tumors in the AAV-Vector or AAV-sgTrp53 injected groups. While the IVIS data suggested no significant change between ICB therapy and PBS treatment groups, anti-PD1 (n = 11) or anti-CTLA4 (n = 11) treatment prolonged overall survival in comparison to mice receiving PBS treatment (n = 15) (Fig. 1B; Supplementary Fig. S1B). We then harvested all liver lobes for genomic sequencing and histological characterization. Endpoint histological sections from vector-treated mice (n = 3) revealed no tumor lesions, while all mTSG-treated mice (n = 37) developed large and heterogenous liver tumors (Fig. 1C).
We processed the tumors for targeted analysis of the predicted sgRNA cut sites using molecular inversion probe (MIP) sequencing (27). Representative variants of insertions and deletions (indels) detected by MIP capture sequencing are shown at the cut sites of B2m sg3 (Fig. 1D), Arid1a sg4 (Fig. 1E), and Kmt2d sg3 (Fig. 1F). We summed the constituent variant frequencies of each sgRNA and charted the mutation landscape associated with each treatment (Fig. 1G). We then calculated the mean variant frequencies for each gene, grouping samples by treatment condition. By comparing the gene mutation frequencies in the different treatment groups, we uncovered genetic perturbations that were comparatively enriched or depleted with anti-PD1 or anti-CTLA4 treatment (Fig. 1G and 2A). Comparing the anti-CTLA4 treated mice with PBS treated mice, the mutation frequencies of B2m, Grlf1, Bcor and Kdm5c were significantly increased, whereas the mutation frequencies of Arid1a were significantly decreased (Fig. 2B). Comparing anti-PD1 treated mice with PBS treated mice, knockout of B2m, Grif1, Vhl, Cdkn1b, or Bcor was correlated with anti-PD1 resistance, while the knockout of Kmt2d, Arid1a, Rnf43 or Atrx was associated with anti-PD1 responsiveness (Fig. 2C).
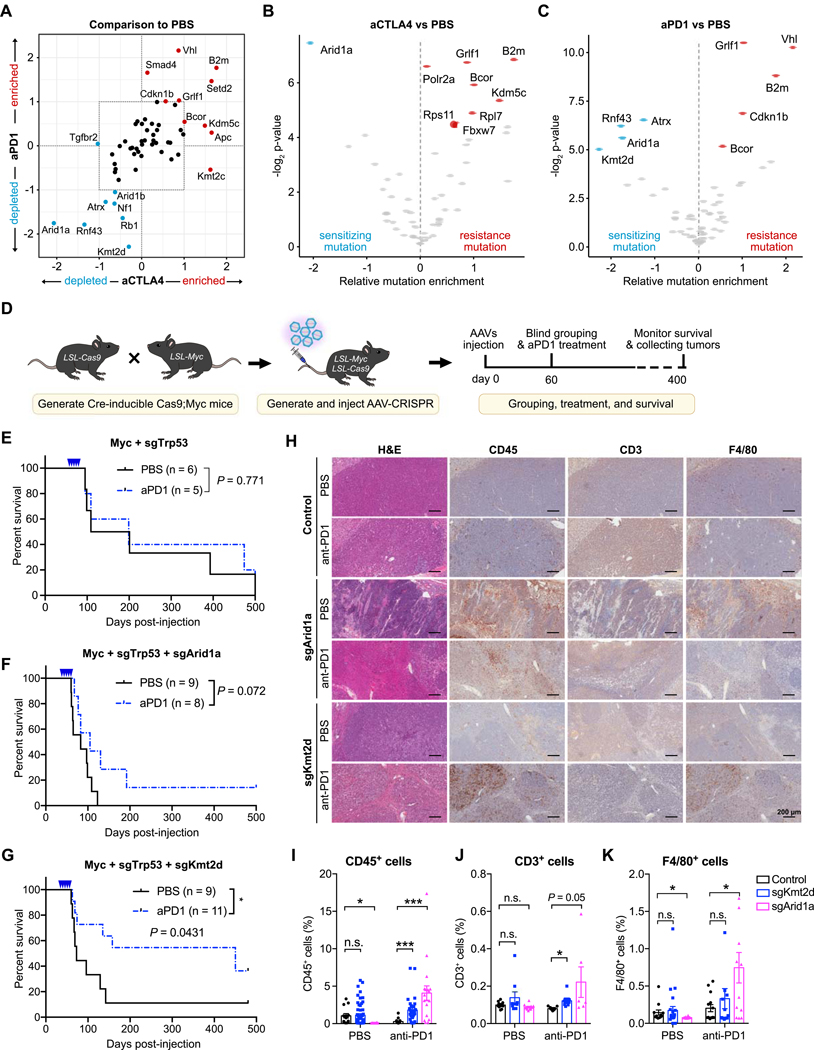
Figure 2. Identification and validation of genetic factors that modulate response to checkpoint immunotherapy.
(A) Scatter plot of average mutation enrichment scores across aCTLA4 or aPD1 treated samples, subtracted by the average score in PBS samples. Negative values indicate relative depletion, while positive values indicate relative enrichment.
(B) Volcano plot comparing the mutation enrichment scores in aCTLA4 vs. PBS treated samples. Negative mutation enrichment scores indicate gene mutations that confer sensitivity to aCTLA4 treatment upon CRISPR mutagenesis, while positive scores indicate gene mutations that confer resistance.
(C) Volcano plot comparing the mutation enrichment scores in aPD1 vs. PBS treated samples. Negative mutation enrichment scores indicate gene mutations that confer sensitivity to aPD1 treatment upon CRISPR mutagenesis, while positive scores indicate gene mutations that confer resistance.
(D) Schematic of experimental design for single gene validation experiments. AAV-CRISPR vectors with a liver-specific Cre expression cassette were intravenously injected into LSL-Cas9; LSL-Myc mice to induce Myc overexpression and Cas9 expression for sgRNA-mediated mutagenesis. Mice were subsequently treated with PBS or aPD1.
(E) Kaplan-Meier survival curves of mice bearing liver tumors with Myc overexpression and Trp53 knockout. PBS (n = 6) and aPD1 (n = 5) treated mice showed no significant survival difference (P = 0.581).
(F) Kaplan-Meier survival curves of mice bearing liver tumors with Myc overexpression, Trp53 knockout, and Arid1a knockout. PBS (n = 9) and aPD1 (n = 8) treated mice showed no significant survival difference (P = 0.072).
(G) Kaplan-Meier survival curves of mice bearing liver tumors with Myc overexpression, Trp53 knockout, and Kmt2d knockout. PBS (n = 9) and aPD1 (n = 11) treated mice showed a significant survival difference (P = 0.0231).
(H) Representative images of H&E, CD45, CD3, and F4/80 staining of liver sections from Myc+sgTrp53, Myc+sgTrp53+sgArid1a, Myc+sgTrp53+sgKmt2d tumors with or without anti-PD1 treatment. Scale bar is 200 μm.
(I-K) Quantification of (I) CD45+ immune cells, (J) CD3+ T cells, or (K) F4/80+ macrophages in liver sections from control, Kmt2d-mutant, or Arid1a-mutant tumors, with or without anti-PD1 treatment. (I) CD45+ cells in different groups. Two-tailed unpaired t-test, CD45+ cells in PBS group: sgKmt2d (n = 57) vs. control (n = 14), P = 0.8672; sgArid1a (n = 16) vs. control (n = 57), P = 0.0012. CD45+ cells in anti-PD1 group: sgKmt2d (n = 31) vs. control (n = 17), P = 0.0008; sgArid1a (n = 18) vs. control (n = 17), P = 0.0005. (J) CD3+ cells in different groups T-test, CD3+ T cells in PBS group: sgKmt2d (n = 9) vs. control (n = 10), P = 0.1988; sgArid1a (n = 11) vs. control (n = 10), P = 0.1373. CD3+ T cells in anti-PD1 group: sgKmt2d (n = 8) vs. control (n = 9), P = 0.0026; sgArid1a (n = 6) vs. control (n = 9), P = 0.050. (K) F4/80+ cells in different groups. T-test, F4/80+ cells in PBS group: sgKmt2d (n = 25) vs. control (n = 12), P = 0.708; sgArid1a (n = 14) vs. control (n = 12), P = 0.0454. CD3+ T cells in anti-PD1 group: sgKmt2d (n = 16) vs. control (n = 14), P = 0.4038; sgArid1a (n = 12) vs. control (n = 14), P = 0.0104. (N represents different IHC staining regions of the slides collected from ≥ 2 mice per treatment group)
Error bars: All data points in this figure are presented as mean ± SEM. Asterisks: * P < 0.05, ** P < 0.01, *** P < 0.001.
See also: Supplementary Fig. S1
Loss-of-function mutations of KMT2D potentiates anti-PD1 checkpoint immunotherapy
From our screen data, Arid1a loss sensitized tumors to both anti-CTLA4 and anti-PD1 treatment, while Kmt2d mutations showed the largest magnitude of sensitization to anti-PD1. To individually validate these findings, we developed and utilized several genetic liver cancer models. We first crossed CAG-LSL-Myc transgenic mice with LSL-Cas9 mice, and then injected these mice with AAVs carrying TBG-Cre and sgTrp53 to drive ectopic Myc expression and Trp53 knockout in the liver (Fig. 2D). To interrogate the effects of concurrent Kmt2d or Arid1a mutations in the setting of Myc overexpression and Trp53 loss, we injected AAVs carrying sgTrp53+sgKmt2d or sgTrp53+sgArid1a into LSL-Myc;LSL-Cas9 mice. As a control, we injected sgTrp53+sgNTC AAVs. Mice bearing Myc+sgTrp53 tumors had a median survival of 155 days, with 50% (3/6) of mice dying within 4 months, and anti-PD1 treatment did not show a significant survival benefit (median survival of 199 days) (Fig. 2E). Additional Arid1a mutations on top of Myc+sgTrp53 resulted in more aggressive tumors, with 100% (9/9) of mice dying within 4 months (median survival of 83 days). Treatment with anti-PD1 marginally prolonged survival (median survival 105 days), with 12.5% (1/8) of mice alive 400 days post-injection (Fig. 2F). Similar results were found using a liver cancer model with Trp53 and Apc knockout as the genetic background, with Arid1a-mutant tumors showing a significant response to anti-PD1 (Supplementary Fig. S1C–E)
For mice bearing Myc+sgTrp53+sgKmt2d tumors, 8/9 (88.9%) of the mice died within 4 months (median survival of 73 days), indicating that KMT2D functions as a tumor suppressor in this context. Strikingly, anti-PD1 therapy prolonged survival of mice bearing Myc+sgTrp53+sgKmt2d tumors, with >50% mice alive at 400 dpi (Fig. 2G). We collected endpoint liver tumors for histology and immunohistochemical characterization of these tumors (Fig. 2H–K), observing that Arid1a- or Kmt2d-mutant liver tumors were more infiltrated with CD45+ immune cells (Fig. 2I) and CD3+ T cells (Fig. 2J), particularly after anti-PD1 therapy. Upon anti-PD1 treatment, F4/80+ macrophages were more abundant in Arid1a-mutant liver tumors, with similar trends in Kmt2d-mutant liver tumors (Fig. 2K). Collectively, these data demonstrate that Arid1a and Kmt2d encode functional tumor suppressors in the liver, and autochthonous liver tumors with mutations in Kmt2d are more likely to respond to anti-PD1 treatment, validating the results from the initial screen.
Kmt2d deficiency sensitizes multiple cancer types to anti-PD1 therapy
To further assess the role of Kmt2d loss on the cancer-immune interactions in liver cancer, we first established a primary tumor cell line from autochthonous Myc+sgTrp53 liver tumors generated in C57BL/6J (B6) mice (MA1L cells). We then transduced these cells with either vector control or sgKmt2d (Supplementary Fig. S2A). As Kmt2d deficiency has previously been implicated in genome instability (30), we cultured MA1L-Vector and MA1L-sgKmt2d cells in vitro for approximately 100 days and then transplanted the cells into mice to investigate the impact of Kmt2d deficiency. MA1L-sgKmt2d cells formed larger tumors in immunocompromised Rag1−/− mice compared to vector control, again indicating the role of KMT2D as a tumor suppressor (Supplementary Fig. S2B). In contrast, in immunocompetent C57BL/6J mice, the tumors formed by Kmt2d mutant cells were eliminated more rapidly compared to the control (Supplementary Fig. S2C). Notably, the MA1L cell line was established from an endpoint liver tumor and therefore had likely accumulated genetic/epigenetic alterations that increased the immunogenicity of the MA1L cells. Additionally, the immunogenicity of Cas9 in the CRISPR system may contribute to the rejection of the MA1L cells by C57BL/6J mice. Nevertheless, these data reaffirm that Kmt2d mutation sensitizes liver tumors to immune rejection.
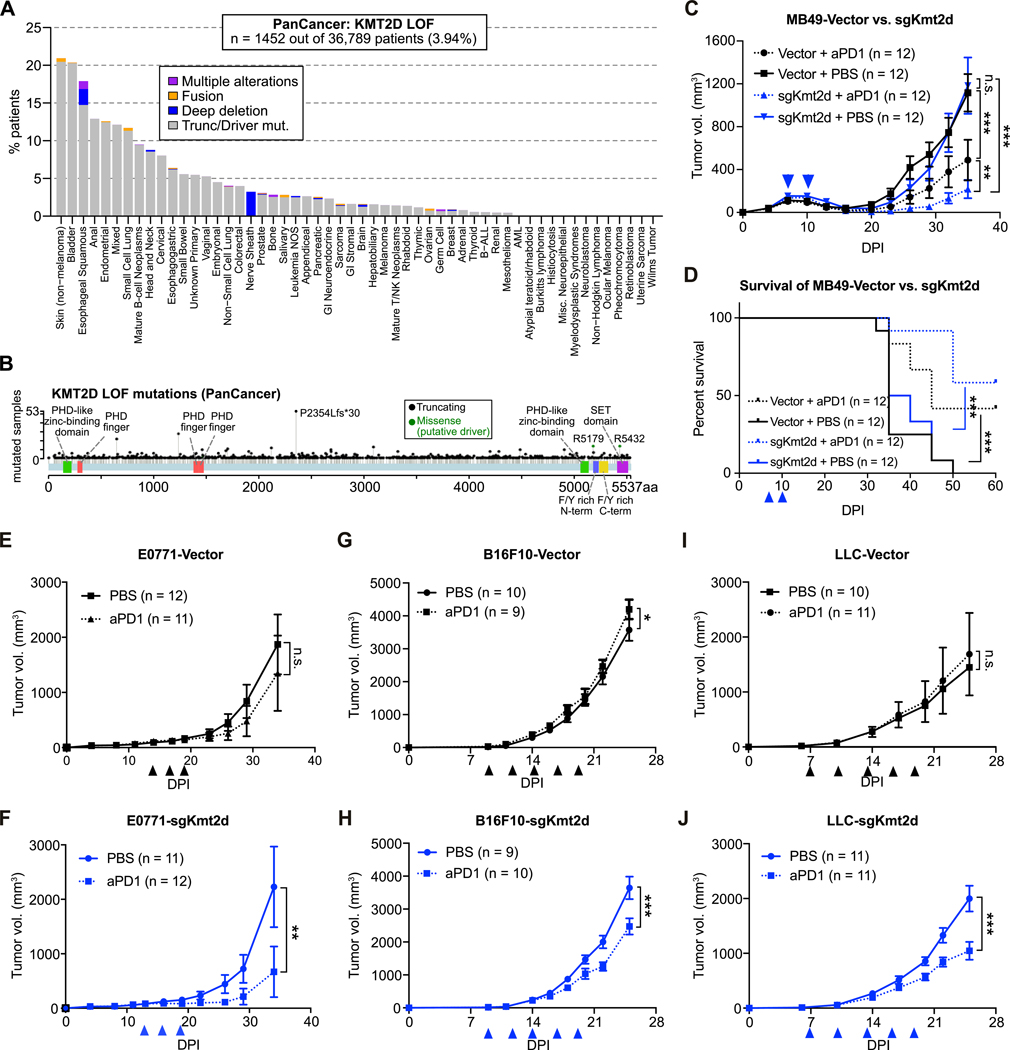
KMT2D is higly mutated in multiple human cancer types, with an average mutation frequency of 4–8% across all cancer patients, and over > 20% of patients with skin cancer and bladder cancer (Fig. 3A; Supplementary Fig. S2D). These mutations are often truncating mutations or putative driver missense mutations (Fig 3B; Supplementary Fig. S2D), supporting the general role of KMT2D as a tumor suppressor gene in humans. Given the prevalence of KMT2D mutations across diverse human cancers, we sought to investigate whether Kmt2d deficiency promotes anti-PD1 responsiveness in other tumor types.
Figure 3. KMT2D loss-of-function mutations are prevalent and associated with improved responses to aPD1 therapy across diverse cancer types.
(A) Bar plot of the percentage of patients with loss-of-function (LOF) or putative driver mutations in KMT2D across multiple cancer types.
(B) Landscape of truncating and putative driver missense mutations in KMT2D across multiple cancer types.
(C) Growth curves of MB49 bladder cancer cells transduced with Vector or sgKmt2d, transplanted into syngeneic C57BL/6J mice. Tumor-bearing mice were treated with either PBS (solid line) or aPD1 (dotted line) at the indicated times (arrowheads). Two-way ANOVA: MB49-Vector, PBS (n = 12) vs aPD1 (n = 12), P < 0.0001; MB49-sgKmt2d, PBS (n = 12) vs aPD1 (n = 12), P < 0.0001; PBS group, MB49-sgKmt2d (n = 12) vs. MB49-Vector (n = 12), P = 0.5988; aPD1 group, MB-sgKmt2d (n = 12) vs. MB49-Vector (n = 12), P = 0.0081.
(D) Kaplan-Meier survival curves of mice bearing MB49 bladder cancer cells transduced with Vector or sgKmt2d that were treated by PBS and aPD1 respectively. Log-rank test: MB49-Vector, aPD1 (n = 12) vs. PBS (n = 12), P = 0.0046; MB49-sgKmt2d, aPD1 (n = 12) vs. PBS (n = 12), P < 0.0001.
(E-F) Growth curves of E0771 triple-negative breast cancer cells transduced with Vector (E) or sgKmt2d (F) in syngeneic C57BL/6J mice. Mice were treated with either PBS (solid line) or aPD1 (dotted line) at the indicated times (arrowheads). Two-way ANOVA: E0771-Vector, PBS (n = 12) vs aPD1 (n = 11), P = 0.3677; E0771-sgKmt2d, PBS (n = 11) vs aPD1 (n = 12), P = 0.005.
(G-H) Growth curves of B16F10 melanoma cells transduced with Vector (G) or sgKmt2d (H), transplanted into syngeneic C57BL/6J mice. Mice were treated with either PBS (solid line) or aPD1 (dotted line) at the indicated times (arrowheads). Two-way ANOVA: B16F10-Vector, PBS (n = 10) vs aPD1 (n = 9), P = 0.012; B16F10-sgKmt2d, PBS (n = 9) vs aPD1 (n = 10), P < 0.0001.
(I-J) Growth curves of Lewis lung cancer (LLC) cells transduced with Vector (I) or sgKmt2d (J), transplanted into syngeneic C57BL/6J mice. Mice were treated with either PBS (solid line) or aPD1 (dotted line) at the indicated times (arrowheads). Two-way ANOVA: LLC-Vector, PBS (n = 10) vs aPD1 (n = 11), P = 0.1867; LLC-sgKmt2d, PBS (n = 11) vs aPD1 (n = 11), P < 0.0001.
Data were collected from 2 independent experiments. Error bars: All data points in this figure are presented as mean ± SEM. Asterisks: * P < 0.05, ** P < 0.01, *** P < 0.001.
See also: Supplementary Fig. S2, S3
As KMT2D is highly mutated in human bladder cancers, we mutated Kmt2d in MB49 bladder cancer cells (MB49-sgKmt2d; Supplementary Fig. S2E) and transplanted the cells into C57BL/6J mice, with vector-transduced cells (MB49-Vector) as a control. When treated at an early stage, both MB49-sgKmt2d and MB49-Vector tumors responded to anti-PD1 treatment (Fig. 3C). However, MB49-sgKmt2d tumors were comparatively more responsive to anti-PD1 therapy (Fig. 3C). Accordingly, anti-PD1 treatment significantly improved overall survival, with MB49-sgKmt2d tumor-bearing mice surviving slightly longer than mice with MB49-Vector tumors (Fig. 3D). Examining the tumor growth kinetics, we noted an early stage of immune elimination of the MB49 tumors around 10 dpi. We therefore investigated the responses of late-stage MB49 tumors to anti-PD1 therapy. When treated at a later timepoint after the initial immune response, MB49-Vector tumors did not respond to anti-PD1 treatment (Supplementary Fig. S2F), but Kmt2d-mutant tumors were still responsive to anti-PD1 treatment (Supplementary Fig. S2G). Anti-PD1 therapy showed a significant survival benefit in mice bearing MB49-sgKmt2d tumors, but not in mice with MB49-Vector tumors (Supplementary Fig. S2H).
Similarly, we mutated Kmt2d in E0771 triple-negative breast cancer cells, B16F10 melanoma cells, and Lewis lung cancer (LLC) cells (Supplementary Fig. S2I), and then transplanted them into C57BL/6J mice. For the orthotopic E0771 model, only the mice bearing Kmt2d-mutant tumors significantly benefited from the anti-PD1 treatment (Fig. 3E and F; Supplementary Fig. S2J and K). Orthotopic tumors formed by vector-transduced B16F10 cells (B16F10-Vector) were resistant to anti-PD1 treatment (Fig. 3G), but the addition of Kmt2d loss sensitized the tumors to anti-PD1 treatment (Fig. 3H). Similarly, subcutaneous Vector-transduced LLC tumors were resistant to anti-PD1 treatment, but Kmt2d-mutant LLC tumors partially responded to anti-PD1 treatment (Fig. 3I and J).
To examine whether Kmt2d loss induces anti-PD1 sensitivity by specifically reshaping the local microenvironment, we utilized a dual-tumor model in which we transplanted LLC-Vector cells in the left flanks and LLC-sgKmt2d cells in the right flanks of individual mice. We found that only the Kmt2d-mutant tumors responded to anti-PD1, anti-PDL1, or anti-CTLA4 treatment (Supplementary Fig. S3A–D), suggesting that Kmt2d loss sensitizes tumors to checkpoint therapy by altering the tumor microenvironment. Together, these results demonstrate that Kmt2d mutation promotes anti-PD1 response in 4 additional tumor types (bladder cancer, triple-negative breast cancer, melanoma, and lung cancer), generalizing the findings from the CRISPR-GEMM liver cancer system (Fig. 2G).
Kmt2d-mutant tumors exhibit enhanced immune infiltration in the tumor microenvironment
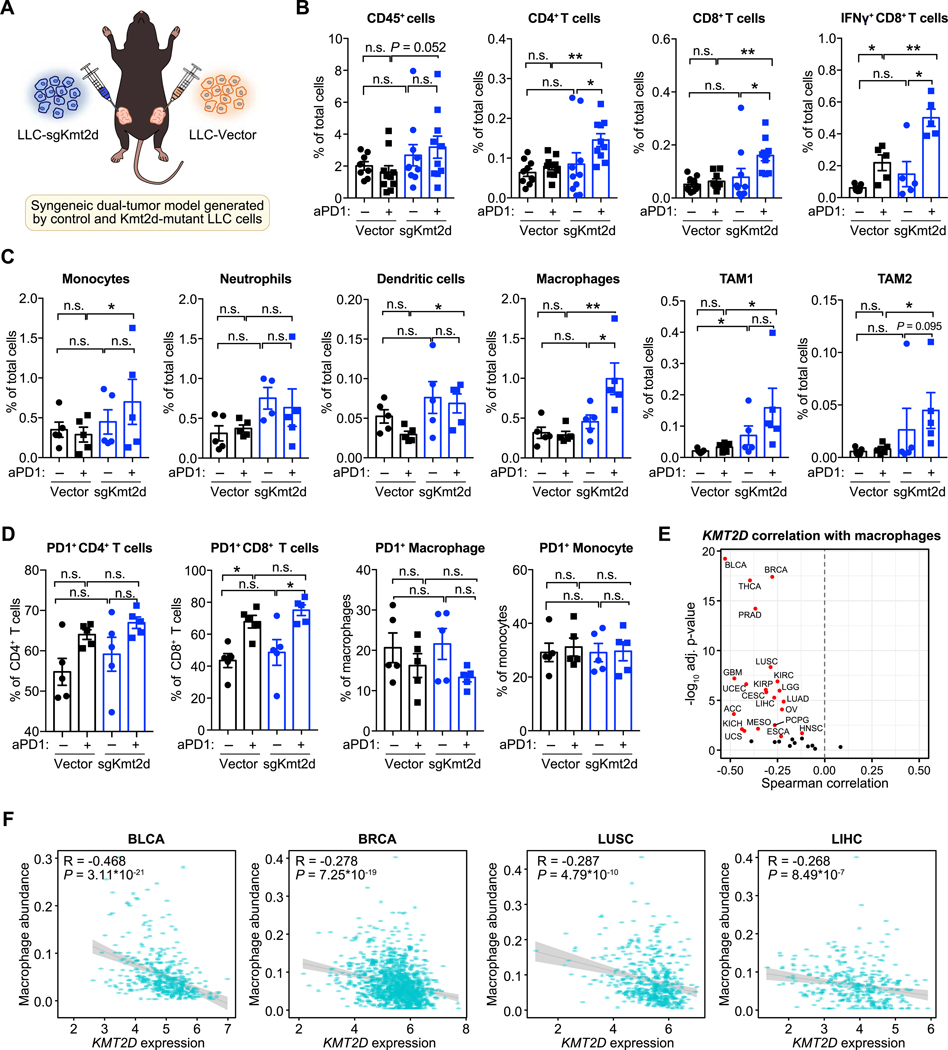
To investigate the mechanisms underlying the enhanced anti-tumor response against Kmt2d-mutant MA1L liver tumors, we analyzed the tumor immune microenvironment by flow cytometry (Supplementary Fig. S4) at day 11 when tumor regression begins. We found that Kmt2d-mutant MA1L tumors had more CD45+ immune cells, CD4+ T cells, and macrophages compared to controls (Supplementary Fig. S5A). CD8+ T cell infiltration also trended towards an increase in Kmt2d-mutant tumors, although not statistically significant. To examine whether Kmt2d knockout consistently promotes immune infiltration, we further analyzed the tumor microenvironment of control or Kmt2d-mutant MB49 bladder cancers. We similarly found increased infiltration of CD45+ immune cells, especially CD8+ T cells, in MB49-sgKmt2d tumors (Supplementary Fig. S5B).
Since the MA1L liver tumors and MB49 bladder tumors were often rejected after anti-PD1 therapy, we used the anti-PD1-resistant LLC model to further investigate the effect of anti-PD1 therapy on Kmt2d-mutant tumors. At 19 days post-induction of the dual-tumor LLC model (Fig. 4A), we analyzed the immune context of tumors formed by LLC-sgKmt2d and LLC-Vector cells, with or without anti-PD1 therapy (Fig. 4B; Supplementary Fig. S6A). We observed that Lewis-sgKmt2d tumors had increased infiltration of CD45+ immune cells compared to Lewis-Vector tumors, particularly after anti-PD1 treatment (Fig. 4B; Supplementary Fig. S6B). In Kmt2d-mutant tumors, we found significantly increased infiltration of T cells, including CD4+ T cells, CD8+ T cells, and IFN-γ+ CD8+ T cells (Fig. 4B), as well as increased antigen-presenting cells, such as dendritic cells and macrophages (Fig. 4C). These changes became apparent after anti-PD1 treatment. No difference was observed in the abundance of neutrophils and regulatory T cells (Fig. 4C; Supplementary Fig. S6A). By further analyzing the polarizations of tumor-infiltrating macrophages, we found the macrophages were dominated by tumor-associated macrophage 1 (TAM1), although the abundances of TAM1 and TAM2 were both increased in Kmt2d-mutant tumors after anti-PD1 treatment (Fig. 4C). Notably, tumor-infiltrating T cells and innate immune populations (i.e. monocytes, neutrophils and macrophages) expressed PD1 on their surface (Fig. 4D; Supplementary Fig. S6A). Similar results were obtained using the single-tumor LLC model (Supplementary Fig. S6B). These data suggest that the enhanced efficacy of anti-PD1 therapy in Kmt2d-mutant tumors may be due to its effects on T cells as well as myeloid cells, in concordance with a recent observation in these cell types (31).
Figure 4. Kmt2d-mutant tumors are associated with increased innate and adaptive immune infiltration.
(A) Schematic of experimental design for generating the syngeneic LLC dual-tumor model. LLC-Vector and LLC-sgKmt2d cells were transplanted into the left and right flanks of C57BL/6J mice, respectively, followed by treatment with PBS or anti-PD1. Tumor-infiltrating immune cells were analyzed by FACS analysis.
(B) Abundance of intratumoral CD45+ pan-immune cells, CD4+ T cells, CD8+ T cells, and IFN-γ+ CD8+ T cells in LLC-Vector or LLC-sgKmt2d tumors, treated with PBS (n ≥ 5) or aPD1 (n ≥ 5). Mann-Whitney test: sgKmt2d + aPD1 vs. Vector + aPD1: CD45+ cells, P = 0.052; CD4+ T cells, P = 0.0029; CD8+ T cells, P = 0.0003; IFN-γ+ CD8+ T cells, P = 0.0079.
(C) The intratumoral abundance of monocytes, neutrophils, dendritic cells, macrophages, TAM1 (tumor-associated macrophage 1) and TAM2 in LLC-Vector or LLC-sgKmt2d tumors, treated with PBS (n = 5) or aPD1 (n = 5). Mann-Whitney test, sgKmt2d + aPD1 vs Vector + aPD1: monocytes, P = 0.4206; neutrophils, P = 0.3095; dendritic cells, P = 0.0159; macrophages, P = 0.0079; TAM1, P = 0.0317; TAM2, P = 0.0159.
(D) PD1-expression in different immune populations present within LLC-Vector or LLC-sgKmt2d tumors, treated with PBS (n = 5) or aPD1 (n = 5). Mann-Whitney test, sgKmt2d + aPD1 vs Vector + aPD1: PD1+CD4+ T cells, P = 0.2222; PD1+CD8+ T cells, P = 0.2222; PD1+ macrophages, P = 0.4206; PD1+monocytes, P = 0.6905.
(E) Volcano plot of the Spearman correlation between KMT2D mRNA expression and macrophage abundance in 33 cancer types. Red dots indicate cancer types in which KMT2D is significantly negatively correlated with macrophage abundance (adjusted P < 0.05).
(F) Scatter plots comparing KMT2D expression and macrophage abundance in TCGA BLCA, BRCA, LUSC, and LIHC cohorts.
Error bars: All data points in this figure are presented as mean ± s.e.m. Asterisks: * P < 0.05, ** P < 0.01, *** P < 0.001.
See also: Supplementary Fig. S4, S5, S6
To assess the relevance of these findings in clinical cohorts, we evaluated the correlation of KMT2D expression and immune infiltration status across multiple human cancers. In The Cancer Genome Atlas (TCGA), KMT2D expression levels are negatively correlated with intratumoral macrophage abundance in 21/33 (63.6%) of human cancer types (Fig. 4E and F). Similarly, we observed that across multiple cancer types, KMT2D expression is negatively correlated with expression of the monocyte-macrophage marker CD14 and the cytotoxic T cell markers GZMB and GZMA (Supplementary Fig. S7A–C). Collectively, these analyses indicate intratumoral macrophage and cytotoxic CD8+ T cell abundance is increased in tumors with low KMT2D levels, substantiating the results from the FACS data.
Kmt2d deficiency leads to elevated DNA damage and mutation burden
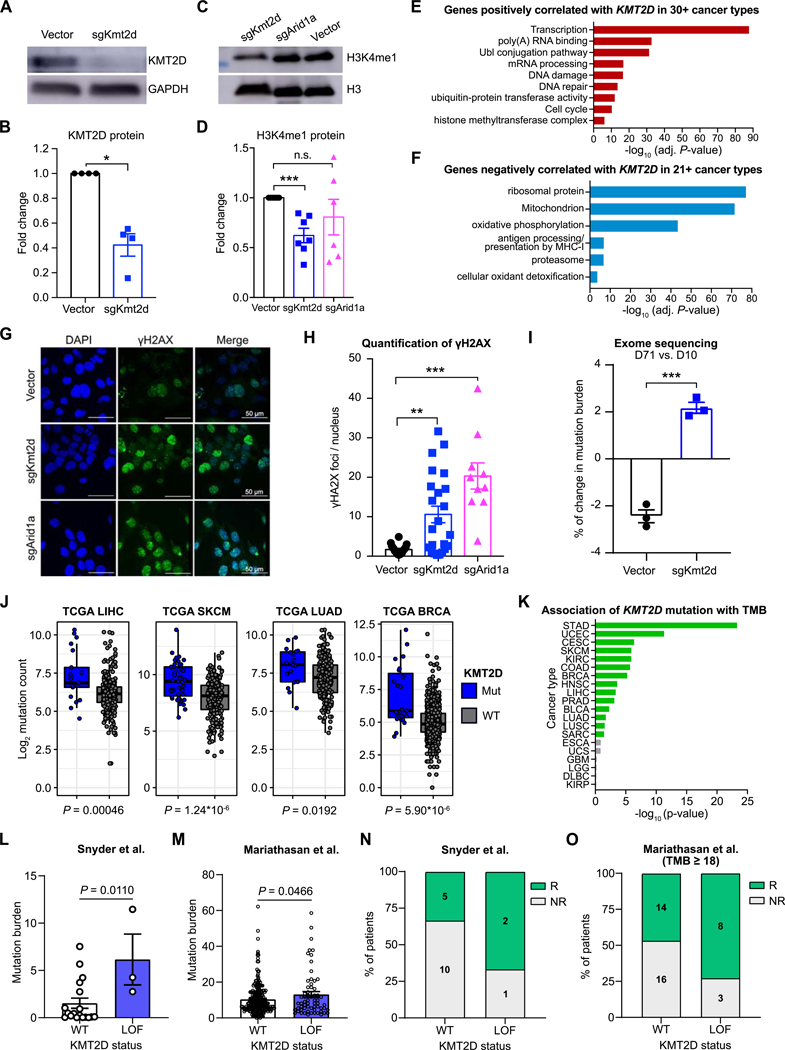
We sought to unravel the mechanisms by which Kmt2d mutation leads to elevated immune infiltration. We first validated that CRISPR targeting of Kmt2d led to a loss of KMT2D protein (Fig. 5A and B), with decreased levels of H3K4me1 but not H3K4me3 (Fig. 5C and D; Supplementary Fig. S7D). To explore the consequences of KMT2D loss, we further analyzed the TCGA data and identified all genes that are significantly correlated with KMT2D expression. To pinpoint associations that are generalizable across multiple cancer types, we selected genes that were concordantly correlated with KMT2D in several independent cohorts (Supplementary Fig. S7E). Genes positively correlated with KMT2D are enriched for transcription, poly(A) RNA binding, Ubl conjugation pathway, mRNA processing, DNA damage and repair, and Ubiquitin-protein transferase activity (Fig. 5E). Genes negatively correlated with KMT2D expression are enriched in ribosomal protein, mitochondrion, oxidative phosphorylation, antigen processing and presentation by MHCI, proteasome, and cellular oxidant detoxification (Fig. 5F). These results indicate that KMT2D has an important role in transcriptional regulation and DNA repair. Indeed, recent studies showed that KMT2D could prevent collisions between transcription and replication machineries (32), and Kmt2d mutation resulted in transcription stress and DNA breaks in replicating regions (30).
Figure 5. Loss of Kmt2d leads to DNA damage and increased mutation burden.
(A) Western blot of KMT2D and GAPDH expression in MA1L cells isolated from the liver tumors of Myc+ Trp53−/− mice, transduced ex vivo with either Vector or sgKmt2d.
(B) Quantification of KMT2D protein, normalized to GAPDH internal control. Mann-Whitney test, Vector (n = 4) vs. sgKmt2d (n = 4), P = 0.0286.
(C) Western blot analysis of H3K4 mono-methylation level (H3K4me1) and total H3 in MA1L cells transduced with Vector, sgKmt2d, or sgArid1a.
(D) Quantification of H3K4me1 levels, normalized to total H3. Mann-Whitney test, Vector (n = 7) vs. sgKmt2d (n = 7), P = 0.0006; Vector (n = 7) vs. sgArid1a (n = 6), P = 0.3065.
(E) DAVID gene ontology analysis of genes positively correlated with KMT2D expression in ≥ 30 cancer types.
(F) DAVID gene ontology analysis of genes negatively correlated with KMT2D expression in ≥ 21 cancer types.
(G) Representative images of γH2AX immunofluorescent staining in MA1L cells transduced with Vector, sgKmt2d or sgArid1a.
(H) Quantification of γH2AX nuclear foci in MA1L cells transduced with Vector, sgKmt2d or sgArid1a. Mann-Whitney test, Vector (n = 18) vs. sgKmt2d (n = 23), P = 0.0022; Vector (n = 18) vs. sgArid1a (n = 10), P < 0.0001.
(I) Exome sequencing analysis of MA1L cells transduced with Vector or sgKmt2d, 10 days (D10) or 71 days (D71) post-transduction. The percentage change in mutation burden was calculated for each condition, comparing D71 (n = 3) to D10 (n = 3). Two-tailed unpaired t-test, P = 0.0003.
(J) Box plots of log2 mutation count in TCGA LIHC, SKCM, LUAD, and BRCA cohorts, grouped by KMT2D mutation status. KMT2D mutant vs wildtype, two-tailed Mann-Whitney test: LIHC, P = 0.00046; SKCM, P = 1.24*10−6; LUAD, P = 0.0192; BRCA, P = 5.90*10-6.
(K) Bar plots detailing the association between KMT2D mutation and total mutation burden (TMB) across cancer types (filtered by cancer types with at least 5 KMT2D-mutant tumors).
(L) Comparison of tumor mutation burden between patients with wildtype or mutant KMT2D in a cohort of bladder cancer patients receiving anti-PD-L1 ICB (Snyder et al.) (34). KMT2D LOF mutant vs wildtype, Mann-Whitney test: P = 0.0110.
(M) Comparison of tumor mutation burden between patients with KMT2D LOF mutations and wildtype KMT2D in the Mariathasan cohort of bladder cancer patients receiving anti-PD-L1 ICB (35). KMT2D LOF mutant vs wildtype, Mann-Whitney test: P = 0.0466.
(N) Relationship between KMT2D status and anti-PD-L1 responses in the Snyder cohort (34).
(O) Relationship between KMT2D status and anti-PD-L1 responses in tumors with mutational burden ≥18 per Mbp in the Mariathasan cohort (35).
Data were collected from ≥ 2 independent experiments. Error bars: All data points in this figure are presented as mean ± SEM.
Asterisks: * P < 0.05, ** P < 0.01, *** P < 0.001.
See also: Supplementary Fig. S7, S8, S9
To assess DNA damage in Kmt2d-mutant and control cells, we used immunofluorescence assays to quantify nuclear γH2AX foci, a marker of unrepaired DNA lesions (Fig. 5G). As a control, we found Arid1a-mutant cells had significantly higher numbers of γH2AX foci compared to the control (Fig. 5H), consistent with the role of ARID1A in mismatch repair (MMR) (33). Notably, we found Kmt2d-mutant cells also had significantly higher levels of γH2AX foci in MA1L liver cancer cells (Fig. 5G and H) and MB49 bladder cancer cells (Supplementary Fig. S8A and B). These differences were not solely due to Cas9-induced double-stranded breaks, since cells transduced with sgKmt2d similarly showed increased γH2AX foci compared to cells transduced with Aavs1 locus (Ppp1r12c)-targeting sgRNA (Supplementary Fig. S8C and D). To confirm that Kmt2d mutation leads to elevated DNA damage, we cultured the Kmt2d-mutant cells and vector control cells in vitro for 71 days and performed whole exome-sequencing. We found that the mutation burden of vector cells decreased over time, whereas the mutation burden of Kmt2d-mutant cells increased (Fig. 5I). This increase was not due to differences in cell proliferation, since cells transduced with sgKmt2d or sgArid1a proliferated at similar rates as Vector-transduced cells (Supplementary Fig. S8E and F).
To examine whether these findings are recapitulated in human cancer, we knocked out KMT2D in H1299 human lung cancer cells (Supplementary Fig. S8G), finding that KMT2D loss led to significantly higher levels of γH2AX and 53BP1 foci (Supplementary Fig. S8H and I). We then analyzed the TCGA datasets and found that the tumor mutation burden (TMB) of KMT2D-mutant tumors was indeed significantly higher than the TMB of KMT2D-wildtype tumors across multiple cancer types (Fig. 5J and K). Since KMT2D is highly mutated in human bladder cancers (Fig. 3A), we further analyzed the correlations of KMT2D mutation with TMB and anti-PD1 responses using two cohorts of bladder cancer patients (34,35). In both cohorts, we found that KMT2D-mutant bladder cancers had significantly higher TMB (Fig. 5L and M), and were more likely to respond to ICB with anti-PD-L1 (Fig. 5N and O), especially in the TMB-high patients (Supplementary Fig. S9A and B). Thus, KMT2D mutation leads to elevated DNA damage and is correlated with higher TMB in multiple types of human cancers, as well as better responses to checkpoint immunotherapy (10).
Kmt2d deficiency reshapes the chromatin accessibility of IFN-γ regulated regions
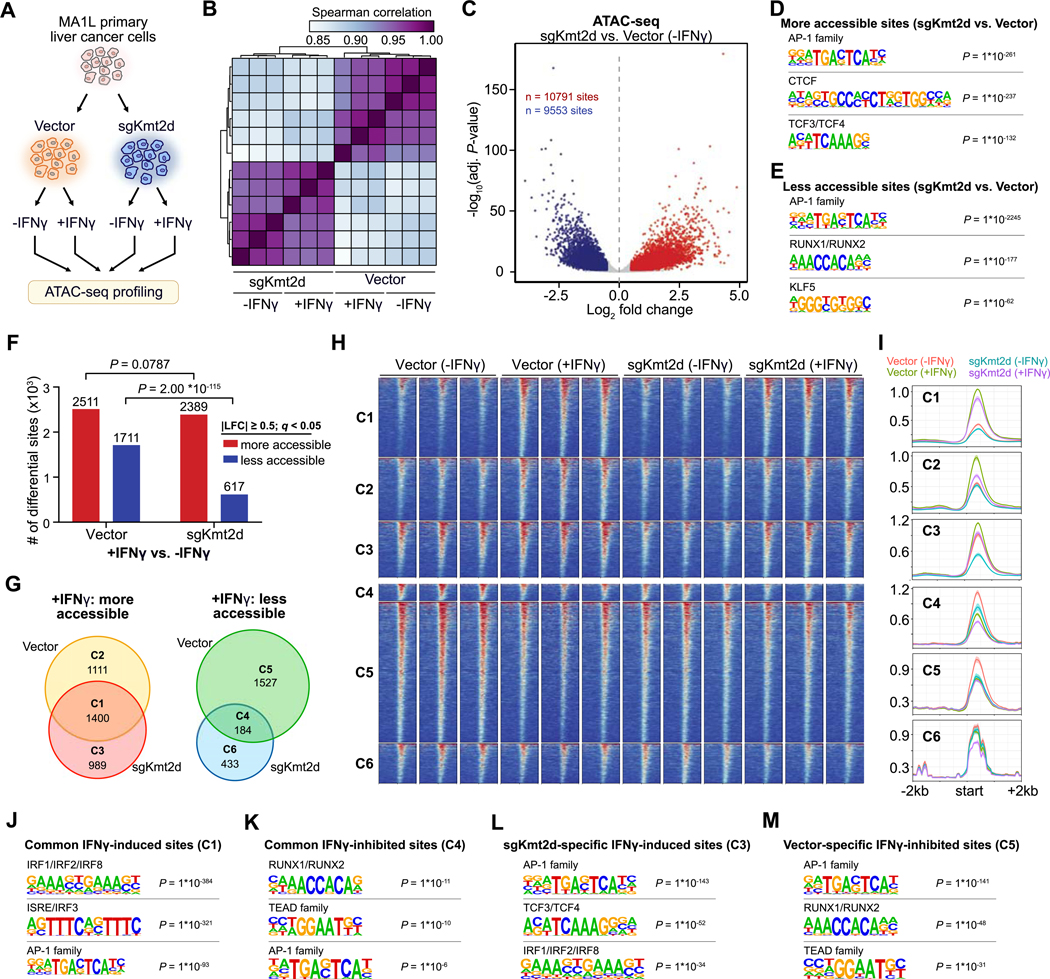
As KMT2D is an epigenetic modifier associated with chromatin regulation and enhancer activation, we performed ATAC-seq to examine the chromatin landscape of Kmt2d-mutant or control MA1L liver tumor cells (Fig. 6A), with or without IFN-γ treatment. Correlation analysis revealed a robust clustering of sgKmt2d cells separately from control cells (Fig. 6B). Comparing Kmt2d-mutant to control cells (without IFN-γ treatment), 10,791 sites were more accessible and 9,553 sites were less accessible (Fig. 6C). Motif analysis of the more accessible sites revealed enrichment for AP-1 family factors, CTCF and TCF3/TCF4 (Fig. 6D), while the less acessible sites were also enriched for AP-1 family factors (Fig. 6E). The common enrichment for AP-1 binding motifs suggests a global rewiring of AP-1-driven programs upon perturbation of Kmt2d.
Figure 6. Kmt2d deficiency remodels the chromatin accessibility of IFN-γ regulated regions.
(A) Schematic of the experimental design. MA1L primary liver cancer cells were transduced with Vector (n = 3) or sgKmt2d (n = 3). Cells were then cultured in the presence or absence of IFN-γ, followed by ATAC-seq profiling.
(B) Heat map detailing the pairwise Spearman correlations of the chromatin accessibility profiles for each of the four conditions.
(C) Volcano plot comparing genome-wide chromatin accessibility in sgKmt2d vs. Vector cells, in the absence of IFN-γ. 10,791 sites were significantly more accessible in sgKmt2d cells, while 9,553 sites were more accessible in Vector cells (adjusted P < 0.05, |log2 fold change| ≥ 0.5).
(D-E) Motif analysis of the genomic regions that are more accessible (d) or less accessible (e) in sgKmt2d vs. Vector cells.
(F) Bar plot detailing the chromatin changes induced by IFN-γ treatment, in Vector or sgKmt2d cells. Red, sites that are more accessible in +IFN-γ vs –IFN-γ conditions, comparing Vector and sgKmt2d cells separately (adjusted P < 0.05, |log2 fold change| ≥ 0.5). Fischer’s exact test, Vector vs. sgKmt2d: number of more accessible sites with IFN-γ treatment, P = 0.0787; less accessible sites with IFN-γ treatment, P = 2.00*10-115.
(G) Venn diagrams of genomic regions that were more accessible (left) or less accessible (right) with IFN-γ treatment, comparing Vector and sgKmt2d cells.
(H) Heat map of normalized ATAC-seq signal across the six clusters defined in (G).
(I) Average signal profiles across the six clusters shown in (H).
(J-K) Motif analysis of the genomic regions that are more (J) or less (K) accessible with IFN-γ stimulation in both Vector and sgKmt2d cells.
(L) Motif analysis of genomic regions that were more accessible with IFN-γ stimulation only in sgKmt2d cells.
(M) Motif analysis of genomic regions that were less accessible with IFN-γ stimulation only in Vector cells.
See also: Supplementary Fig. S9
Upon IFN-γ treatment, we observed large changes in chromatin accesibility. Interestingly, significantly fewer sites became less accessible in sgKmt2d cells compared to Vector cells after IFN-γ treatment, whereas a similar number of sites became more accessible (Fig. 6F). These differentially accessible sites could be broadly classified into 6 clusters (Fig. 6G). Sites in Cluster 3 (989 sites) were less accessible in sgKmt2d cells compared to control cells at baseline prior to IFNγ, but become more accessible after IFN-γ treatment to match the levels in control cells (Fig. 6G–I). Conversely, sites in Cluster 5 were more acessible in control cells compared to sgKmt2d cells prior to IFN-γ treatment, but became less accessible after IFN-γ treatment to a level similar to sgKmt2d cells. These findings indicate that IFN-γ stimulation converges the chromatin landscapes of wildtype and Kmt2d-mutant cells (Supplementary Fig. S9C). Thus, Kmt2d deficiency systematically reshapes the chromatin accessibility of IFN-γ-induced genes at baseline, and IFN-γ stimulation partially normalizes these differences. Motif analysis reaffirmed the systematic rewiring of AP-1 family factors in Kmt2d-mutant cells (Fig. 6J–M), further demonstrating that these alterations also influence chromatin changes in response to IFN-γ treatment.
Kmt2d deficiency remodels the transcriptome, leading to altered chemokine profiles in vitro and in vivo
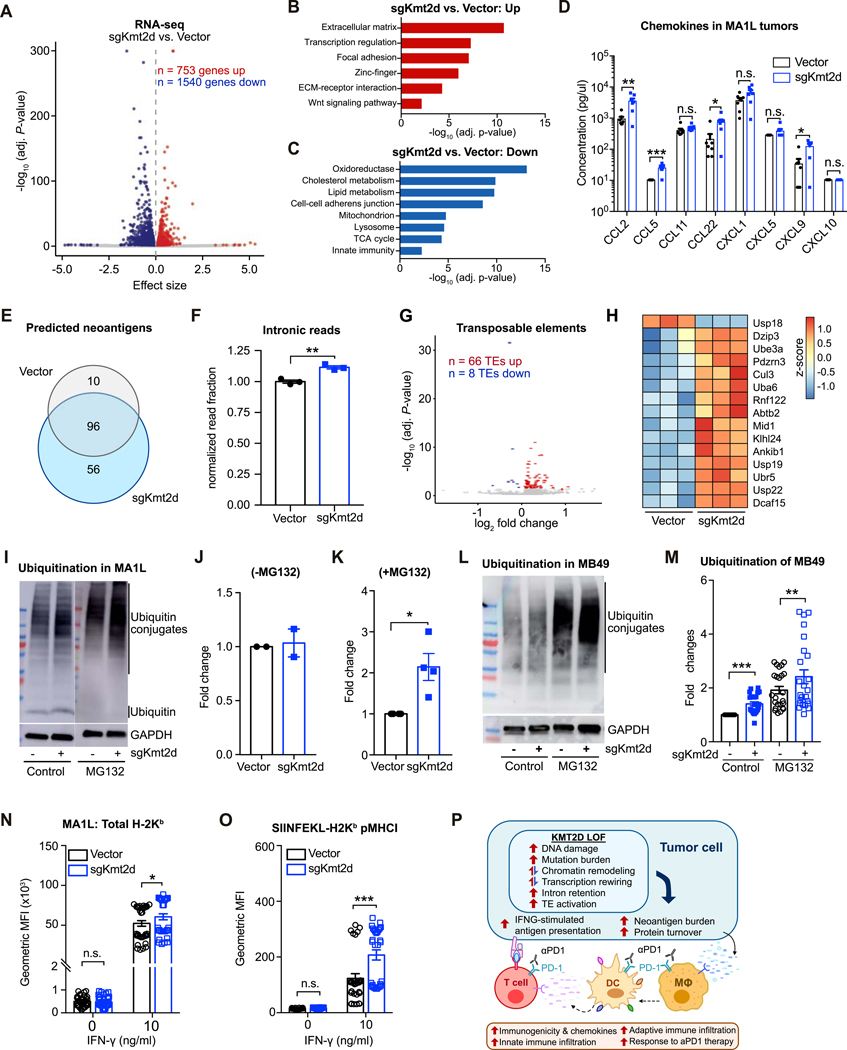
The association of KMT2D with transcription regulation and DNA damage (Fig. 5; Supplementary Fig. S8), along with the broad alterations in chromatin accessibility (Fig. 6), suggest that Kmt2d loss would lead to systematic transcriptional remodeling. To study the transcriptomic changes caused by Kmt2d mutation, we performed RNA-seq on the MA1L liver tumor cells. Comparing to vector controls, we found that 753 genes were upregulated while 1540 genes were downregulated in Kmt2d-mutant cells (Fig. 7A). As the ATAC-seq analyses had pointed to a rewiring of AP-1 family factors, we investigated the expression of different transcription factors that constitute AP-1 dimers. In MA1L-sgKmt2d cells, we observed upregulation of Fosl2, Mafb, and Maf (Supplementary Fig. S9D), and downregulation of Fosl1, Maff, and Atf3 (Supplementary Fig. S9E). To explore the potential downstream consequences of increasing the relative abundance of FOSL2 within AP-1 dimers, we examined FOSL2 ChIP-seq data from HepG2 human liver cancer cells. We found that FOSL2 binds near important regulators of tumor-immune interactions, including IFNGR1, IFNGR2, JAK1, and JAK2 (Supplementary Fig. S9F).
Figure 7. Pleiotropic effects of Kmt2d deficiency on transcriptional regulation, protein turnover, and antigen presentation.
(A) Volcano plot of RNA-seq data, comparing MA1L cells transduced with sgKmt2d (n = 3) vs. Vector (n = 3). 753 genes were significantly upregulated, while 1540 genes were downregulated (adjusted P < 0.05).
(B-C) DAVID gene ontology analysis of genes significantly upregulated (B) or downregulated (C) with Kmt2d deficiency.
(D) Chemokines in the tumors formed by MA1L-Vector and MA1L-sgKmt2d were profiled using the LEGENDplex™ Mouse Proinflammatory Chemokine Panel (13-plex). T-test between MA1L-sgKmt2d (n = 7) vs. MA1L-Vector (n = 7): CCL2, P = 0.0072; CCL5, P = 0.0003; CCL11, P = 0.1961; CCL22, P = 0.0202; CXCL1, P = 0.0948; CXCL5, P = 0.1322; CXCL9, P = 0.0105.
(E) Venn diagram of predicted neoantigens in Vector vs. sgKmt2d transduced cells.
(F) Relative proportion of RNA-seq reads mapping to intronic regions, comparing Vector (n = 3) vs. sgKmt2d (n = 3) transduced cells. Two-tailed unpaired t-test, P = 0.0033.
(G) Volcano plot of transposable element (TE) expression profiles by RNA-seq, comparing MA1L cells transduced with sgKmt2d (n = 3) vs. Vector (n = 3). 66 TEs were significantly upregulated, while 8 TEs were downregulated (adjusted P < 0.05).
(H) Heat map of top differentially expressed genes involved in protein turnover, shown as z-scores.
(I) Western blot analysis of ubiquitin (conjugated or free form) and GAPDH in Vector or sgKmt2d-transduced MA1L liver cancer cells, with (+MG132, right panel) or without (-MG132, left panel) proteasome inhibitor treatment.
(J) Quantification of ubiquitin conjugates in the absence of proteasome inhibition, normalized to Vector.
(K) Quantification of ubiquitin conjugates in the presence of proteasome inhibition, normalized to Vector. Paired t-test, sgKmt2d (n = 4) vs. Vector (n = 4), P = 0.0403.
(L) Western blot analysis of ubiquitin conjugates and GAPDH in Vector or sgKmt2d transduced MB49 bladder cancer cells, with or without the addition of proteasome inhibitor MG132.
(M) Quantification of ubiquitin conjugates in Kmt2d-mutant MB49 bladder cancer cells with or without MG132, normalized to the vector control. Two-tailed paired t-test, without MG132: sgKmt2d (n = 24) vs. Vector (n = 24), P < 0.0001; with MG132: sgKmt2d (n = 24) vs. Vector (n = 24), P = 0.0054.
(N) Flow cytometry analysis of total H-2Kb expression levels in Vector vs. sgKmt2d transduced cells, cultured in 0 ng/ml or 10 ng/ml IFN-γ. Two-tailed unpaired t-test, Vector (n = 36) vs. sgKmt2d (n = 36): 0 ng/ml IFN-γ, P = 0.9931; 10 ng/ml IFN-γ, P = 0.0250.
(O) Flow cytometry analysis of SIINFEKL-H-2Kb peptide-MHC-I complexes in Vector vs. sgKmt2d transduced cells, cultured in 0 ng/ml or 10 ng/ml IFN-γ. Two-tailed unpaired t-test, Vector (n = 30) vs. sgKmt2d (n = 30): 0 ng/ml IFN-γ, P = 0.9109; 10 ng/ml IFN-γ, P < 0.0001. Data of (N-O) were collected from 4 independent experiments
(P) Schematic summarizing the pleiotropic consequences of Kmt2d deficiency on tumor cell-intrinsic properties, leading to increased immune infiltration and potentiating response to aPD1 immunotherapy.
Error bars: All data points in this figure are presented as mean ± SEM. Asterisks: * P < 0.05, ** P < 0.01, *** P < 0.001.
See also: Supplementary Fig. S10 and S11
Analysis of the upregulated genes in Kmt2d-mutant cells revealed multiple enriched categories, including extracellular matrix, transcription regulation, focal adhesion, zinc-finger, ECM-receptor interaction and Wnt signaling (Fig. 7B). Conversely, Kmt2d-mutant cells showed downregulation of oxidoreductase genes and multiple metabolic pathways, including cholesterol metabolism, TCA cycle, and lipid metabolism, as well as mitochondrion, lysosome, and innate immunity (Fig. 7C).
Chemokines (Cxcl1, Cxcl5, Cxcl15) related to neutrophil recruitment were upregulated in Kmt2d-mutant cells compared to the vector control (Supplementary Fig. S10A and B). qPCR validation experiments confirmed the upregulation of Cxcl1 and Cxcl15 mRNA upon Kmt2d loss in both MA1L liver cancer cells and MB49 bladder cancer cells (Supplementary Fig. S10B and C). We then investigated whether these in vitro transcriptional changes were reflected in vivo. When profiling the chemokines in MA1L-derived tumors, we detected significantly higher levels of CCL2, CCL5, CCL22 and CXCL9 protein in MA1L-sgKmt2d tumors, but similar levels of CXCL1 and CXCL5 protein (Fig. 7D). These chemokine changes were notably distinct from the chemokine profiles of in vitro cultured MA1L cells (Supplementary Fig. S10D), suggesting that the upregulation of CCL5, CCL22 and CXCL9 in MA1L-sgKmt2d tumors in vivo was likely contributed by other immune cells that were recruited to the tumors. Thus, the increase of these chemokines may explain the increased recruitment of antigen-presenting cells and T cells in Kmt2d-mutant liver tumors (Fig. S5). However, we did not detect significant changes of these chemokines in MB49 tumors (Supplementary Fig. S10E and F) or LLC tumors (Supplementary Fig. S10G–I). These differences could be caused by a multitude of complex factors, including differences in tumor stage, the cell types involved, and their genetic backgrounds.
Kmt2d deficiency causes intron retention and activation of transposable elements
We next used the RNA-seq data to predict the neoantigens in Kmt2d-mutant and control cells. We found that Kmt2d-mutant and control cells shared 96 predicted neoantigens. Notably, Kmt2d-mutant cells have an additional 56 predicted neoantigens that were not predicted in the control cells, whereas the control cells have 10 additional predicted neoantigens (Fig. 7E). In addition to neoantigens encoded within the canonical proteome, aberrant ribosomal products and alleged noncoding regions can serve as a major source of tumor antigens (36–39). A previous study suggested that Kmt2d mutation leads to transcriptional stress (30), and our analysis of the TCGA implicated KMT2D in transcription regulation and mRNA processing (Fig. 5E). We therefore assessed whether Kmt2d mutation affects RNA splicing and transcript quality by examining intronic retention rates in mRNA. Kmt2d-mutant cells had significantly increased intronic retention in the mRNA, with ~12% more intronic reads than vector control cells (Fig. 7F). This is consistent with a role for KMT2D in transcription regulation and mRNA processing. As many transposable elements (TEs) such as endogenous retroviruses are often contained within introns, we then assessed the expression of TEs in Kmt2d-mutant vs control cells. We observed that 66 TEs were significantly upregulated in Kmt2d-mutant cells, while only 8 TEs were downregulated (Fig. 7G). These data suggested that Kmt2d mutation leads to transcriptional stress and dysregulated RNA splicing, leading to intron retention and heightened expression of TEs. In conjunction with the increased DNA damage and mutation burden upon KMT2D loss, these findings illuminate several sources of potential neoantigens in Kmt2d-mutant cells.
Kmt2d-mutant cells exhibit increased proteasomal degradation and IFN-γ-stimulated antigen presentation
In order to generate antigenic peptides, coding transcripts must first be translated and the resultant proteins ubiquitinated for proteasome-mediated degradation. A number of genes involved in ubiquitination were transcriptionally upregulated in Kmt2d-mutant cells compared to vector control (Fig. 7H). To examine whether the levels of ubiquitinated proteins destined for proteasomal degradation are higher in Kmt2d-mutant cells, we performed immunoblot assays of ubiquitinated proteins with or without MG132, an inhibitor of proteasome degradation (Fig. 7I–M). Without MG132, Kmt2d-mutant cells had either higher or comparable levels of ubiquitinated proteins compared to control (Fig. 7I, J, L,and M; Supplementary Fig. S11A and B). When proteasome degradation was inhibited by MG132, we detected significantly higher levels of ubiquitinated proteins in Kmt2d-mutant MA1L and Kmt2d-mutant MB49 cancer cells compared to the corresponding controls (Fig. 7K and M), though we did not see the same trend in LLC cells (Supplementary Fig. S11A and B). Interestingly, when treated by IFN-γ, both Kmt2d-mutant and control MB49 cells showed higher levels of ubiquitinated proteins (Supplementary Fig. S11C and D). These results indicated that Kmt2d-mutant cells generated more proteins which were ubiquitinated and subjected to proteasomal degradation. This finding is further supported in patient cohorts, as proteasome-related genes were enriched among the genes negatively correlated with KMT2D expression in the TCGA (Fig. 5F).
Following protein ubiquitination and proteasomal degradation, the resultant peptides must be loaded onto major histocompatibility complex I (MHCI) in order to be presented. However, the RNA-seq data revealed that several genes in the MHCI family were downregulated in Kmt2d-mutant MA1L cells compared to the vector control (Fig. S11E), potentially dampening the presentation of potential antigens. Analyzing the levels of cell surface MHCI by flow cytometry, we found that the baseline levels of surface H2-Kb on Kmt2d-mutant cells were comparable to that of vector control (Fig. 7N; Supplementary Fig. S11F). When stimulated by IFN-γ, the surface levels of total H2-Kb were significantly increased and Kmt2d mutant cells exhibited even higher levels of H2-Kb than the control cells (Fig. 7N; Supplementary Fig. S11F). To explicitly test whether tumor antigens would be more efficiently presented in Kmt2d-mutant cells, we transduced the MA1L tumor cells with a phosphoglycerate kinase (PGK) promoter driving ovalbumin (OVA) to examine the presentation levels of H2-Kb-SIINFEKL. We did not observe any difference in the levels of surface H2-Kb-SIINFEKL in Kmt2d-mutant and vector control cells without IFN-γ treatment (Fig. 7O). However, Kmt2d-mutant cells had significantly higher levels of H2-Kb-SIINFEKL than vector control cells when stimulated by IFN-γ (Fig. 7O), indicating that Kmt2d-mutant cells respond strongly to IFN-γ treatment by upregulating antigen presentation.
Taken together, these results demonstrate that Kmt2d mutation in tumor cells may lead to higher levels of neoantigens by causing DNA damage, increasing mutation burden, inducing intronic retention, and activating expression of TEs. Furthermore, Kmt2d mutant cells are characterized by increased proteasomal degradation, as well as increased IFN-γ-stimulated antigen presentation. As a consequence, Kmt2d-mutant tumors exhibit elevated infiltration of PD1+ T cells and macrophages, the latter of which may further amplify the anti-tumor effect of anti-PD1 therapy by activating the adaptive immune system. As a histone methyltransferase, KMT2D likely has pleiotropic effects on tumors; at least in the context of the experiments shown here, a plausible mechanistic explanation is that multiple pathways perturbed by Kmt2d deficiency converge to potentiate response to anti-PD1 immunotherapy (Fig. 7P).
Discussion
In the present study, we performed a mutagenesis screen in a CRISPR-GEMM liver tumor model to pinpoint genetic modulators of immunotherapy response. Distinct from previous studies using in vitro co-culture and tumor transplantation models, our screening system utilizes an autochthonous tumor model that preserves the native microenvironmental context. Here, we systematically mapped the fitness of diverse mutations under immunotherapy treatments, demonstrating the power of this platform for interrogating causal relationships of specific mutations and response to ICB.
Identifying the molecular features that dictate response to immunotherapy has the potential to provide valuable guidance to clinicians. We found that mutations in the H3K4 methyltransferase Kmt2d potentiate response to anti-PD1 therapy in diverse cancer types. Of note, the heightened anti-PD1 response and elevated DNA damage caused by Kmt2d loss does not appear to be dependent on mutant p53. Among the tumor cell lines used in this study, E0771, B16F10 and MA1L cells are p53-deficient, while LLC and MB49 cells are p53-competent. In addition, a prior study that demonstrated that Kmt2d deficiency leads to DNA damage and genomic instability was performed using Trp53-wildtype mouse MEF cells (30). As KMT2D is a tumor suppressor gene that is recurrently mutated across multiple human cancers, the identification of KMT2D deficiency as a predictor of anti-PD1 therapy may have important implications for patient stratification and clinical decision-making.
Our study revealed that Kmt2d-mutant cancer cells exhibited an elevated level of DNA damage and higher mutation burden, and we further corroborated these findings in patient tumor datasets. These findings are consistent with the reported role of KMT2D in genome stability, as KMT2C- and KMT2D- dependent H3K4 methylation at replication forks was found to be involved in replication stress (40). We also found that Kmt2d deficiency led to compromised RNA splicing and activation of TEs. Intron retention and activation of TEs can potentially result in the presentation of immunogenic antigens, and we observed that Kmt2d-mutant cells exhibit increased protein ubiquitination, indicating increased proteasomal degradation. Given the increased mutation burden and aberrant transcription of Kmt2d-mutant cells, we speculate that upregulation of proteasome activity may be a compensatory response to the production of abnormal proteins in these cells. Importantly, the resultant increase in proteasomal degradation is further associated with increased IFNγ-stimulated antigen presentation in these cells, thus providing an explanation for the enhanced sensitivity to anti-PD1 therapy in Kmt2d-mutant tumors.
Interestingly, when we re-challenged mice that had successfully rejected Kmt2d-mutant tumors with either Kmt2d-mutant or wildtype tumor cells, both types of tumors were rejected within 2 weeks (Supplementary Fig. S11G). Although the Kmt2d-mutant cells had accumulated more mutations, it is worth emphasizing that the Kmt2d-mutant and wildtype cells nevertheless share most of their mutations by virtue of their common background, and tumor rejection is often mediated by multifaceted immune responses against multiple antigens. Additionally, Kmt2d-mutant tumors display higher levels of both unique and shared antigens after anti-PD1 treatment, thereby promoting the development of immune memory against both unique and shared tumor antigens. We also note that the CRISPR components may have some degree of immunogenicity, thus contributing to the rejection of these cells by B6 mice; nevertheless, all experiments were conducted in parallel with mutant (gene targeting sgRNAs) groups directly compared to controls (vector or non-targeting guide RNA).
We further found that Kmt2d-mutant cells had increased levels of myeloid-recruiting cytokines Cxcl1 and Cxcl5 at both the RNA and protein level. However, when profiling the chemokines in primary tumors derived from these cells, we instead detected increased CCL2, CCL5, CCL22 and CXCL9 levels in Kmt2d-mutant tumors (Supplementary Fig. S11C), suggesting that the upregulation of these chemokines was due to host immune cells recruited to the tumors. These chemokines would promote the infiltration of antigen-presenting cells and T cells into the tumors. Indeed, Kmt2d-mutant tumors had significantly increased infiltration of antigen-presenting cells, CD4 and CD8 T cells, which all expressed high levels of PD1. Recent work has demonstrated that PD1 blockade promotes anti-tumor immunity not only through its action on T cells, but also by leveraging myeloid-derived innate immune cells towards anti-tumor function (31,41). The elevated immune infiltration in Kmt2d-mutant tumors can therefore be explained by the elevated antigenicity of Kmt2d-mutant cells as well as elevated myeloid cell recruitment. However, a limitation of these data is that certain validation experiments were performed using subcutaneous transplantation models that may not accurately reflect the tumor microenvironment of the cancer’s origin.
In summary, these data collectively demonstrate that Kmt2d loss sensitizes diverse tumor types to checkpoint blockade immunotherapy. This study showcases the power of CRISPR-GEMM models for interrogating complex molecular landscapes in native tumor microenvironments, enabling the dissection of immunotherapeutic responses. Given the prevalence of KMT2D mutations in diverse cancer types, our study could help identify a sizeable patient subpopulation that may have higher chances of being sensitive to ICB therapies such as PD1 checkpoint blockade.
Methods
Institutional Approval.
This study has received institutional regulatory approval. All recombinant DNA and biosafety work was performed under the guidelines of Yale Environment, Health and Safety (EHS) Committee with an approved protocol (Chen-rDNA-15–45; Chen-rDNA-18–45). All animal work was performed under the guidelines of Yale University Institutional Animal Care and Use Committee (IACUC) with approved protocols (Chen-2015–20068; Chen-2018–20068). All human sample work was performed under the guidelines of Yale University Institutional Review Board (IRB) with an approved protocol (HIC#2000020784).
AAV-CRISPR vector and mTSG library cloning.
The AAV-CRISPR vector was designed to express Cre recombinase under a liver-specific TBG promoter. Each vector has two sgRNA expression cassettes, with one of them encoding an sgRNA targeting Trp53, and the other as an open sgRNA expression cassette (double SapI sites for sgRNA cloning). We also designed a liver-specific AAV-CRISPR vector with only one sgRNA expression cassette as a control to study the impact of Trp53 knockout. The mTSG library was generated as previously described (26,27), with over 100x coverage to ensure proper representation of the library.
Production and purification of AAVs carrying mTSG library or individual sgRNA.
8.7 μg of AAV9 serotype plasmid, 10.4 μg of pDF6 helper plasmid, and 5.2 μg of AAV expression plasmid, were added into 450 μl Opti-MEM and mixed well, and then complexed with PEI, incubating at room temperature for 10–15 min before adding them drop-wise into HEK293FT cells at 80–90% confluency. 48–72 h post transfection, the transfected cells were collected. AAVs were purified using chloroform extraction and titrated by qPCR assay (see Supplementary Methods).
Intravenous (i.v.) administration of AAVs for liver transduction.
Rosa26-LSL-Cas9–2A-EGFP (LSL-Cas9) mice were bred with C57BL/6J mice, FVB.129S6(B6)-Gt(ROSA)26Sortm1(Luc)Kael/J mice (LSL-Luc), or C57BL/6N-Gt(ROSA)26Sortm13(CAG-MYC,-CD2*)Rsky/J mice (LSL-Myc for short). Mixed gender (randomized males and females) 8–12 week old mice were used for experiments. For IV injection of AAVs, the mice were restrained in a rodent restrainer (Braintree Scientific). Tails were sterilized by 70% ethanol, and 100–200 μl of concentrated AAVs (~1–2*1011 GCs in total) were injected per mouse. All the mice survived the procedure.
Bioluminescent imaging using IVIS.
After AAV injection, mice were imaged by IVIS each month. Briefly, mice were anesthetized by isofluorane, and then 100–150 μl of 30 mg/ml firefly D-Luciferin potassium salt were intraperitoneally injected with ~ 150 mg/kg body weight. 10–15 minutes after injection, the mice were imaged for in vivo tumor growth using an IVIS machine (PerkinElmer). Relative tumor burden was quantified using LivingImage software (PerkinElmer).
Survival analysis.
We observed that the LSL-Cas9 mice receiving AAV-mTSG i.v. injections rapidly deteriorated in their body condition scores (due to tumor development in most cases). Mice with body condition score (BCS) < 2 were euthanized and the euthanasia date was recorded as the last survival date. Survival data was analyzed by standard Kaplan-Meier method, using Graphpad Prism. Statistical significance was assessed by log-rank test. Mice euthanized early in a healthy state were excluded from calculation of survival percentages.
Genomic DNA extraction from cells and mouse tissues.
The gDNA from frozen ground tissue were purified using DNeasy Blood & Tissue Kits (Qiagen), or standard DNA extraction protocol (see Supplementary Methods). The concentration was measured using a Nanodrop (Thermo Scientific).
Validation using CAG-LSL-Myc transgenic mice plus Trp53 knockout as a tumorigenic background.
Rosa26-LSL-Cas9–2A-EGFP knock-in mice were bred with C57BL/6N-Gt(ROSA)26Sortm13(CAG-MYC,-CD2*)Rsky/J mice (LSL-Myc for short) to obtain LSL-Myc; LSL-Cas9 mice. Mixed gender mice of 7–12 weeks old were used for experiments. Autochthonous liver tumors with Myc overexpression and mutant Trp53 were induced by injecting 1–2*1011 GCs of TBG-Cre AAVs carrying sgTrp53+sgNTC into immunocompetent LSL-Myc;LSL-Cas9 mice. Autochthonous liver tumors with additional Arid1a or Kmt2d mutations were induced by injecting 1–2 *1011 GCs of AAVs carrying sgTrp53 + sgArid1a or sgTrp53 + sgKmt2d into LSL-Myc; LSL-Cas9 mice. Liver tumorigenesis was detectable 60 days after AAV injection, at which point the mice were randomly assigned into 2 groups for 200 μg anti-PD1 treatment or PBS.
Histology and immunohistochemistry.
Liver tumors were collected and fixed in 10% neutral formalin for 2–5 days, then transferred into 70% ethanol. Haematoxylin and eosin (H&E) staining or immunohistochemical staining of CD45, CD3, F4/80, or Cytokeratin pan-cytokeratin were performed on 3–5 μm tissue sections using standard procedures at Yale Pathology Core Facility. To quantify CD45, CD3, and F4/80 positive cells, the slides of different regions of tumor samples were quantitatively scored using the IHC profiler in the ImageJ software (42), and only the percentage distribution of high positive was regarded as positive staining.
Cell lines
HEK293FT cells were purchased from ThermoFisher (Catalog# R70007). E0771 mouse triple-negative breast cancer cells were purchased from CH3 (Catalog# 940001). B16F10 mouse melanoma cells (Catalog# CRL-6475), mouse lewis lung cancer carcinoma cells (Catalog# CRL-1642), and H1299 human lung carcinoma (non-small cell lung cancer) cells (Catalog# CRL-5803) were purchased from ATCC. MB49 mouse bladder cancer carcinoma cells were purchased from Sigma-Aldrich (Catalog# SCC148). MA1L and MA1NC cells were established from autochthonous liver tumors with Myc overexpression and Trp53 knockout mutation generated by the i.v. injection of sgTrp53-targeted AAVs into B6 background LSL-Myc;LSL-Cas9 mice. The cells tested negative for mycoplasma contamination. All the purchased cell lines have been authenticated by the original vendors. All cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin, in a CO2 cell incubator 37°C.
Establishing tumor cell lines from autochthonous liver tumors.
Autochthonous liver tumors with Myc overexpression and Trp53 knockout mutation were generated by the i.v. injection of Trp53-targeted AAVs into LSL-Myc;LSL-Cas9 mice. At the survival endpoint, the liver tumors were isolated and made into single-cell suspension by digestion with collagenase IV after mincing into small pieces and passing through 40 μm cell restrainer. The cells were then cultured in DMEM supplemented with 10% FBS and 1% Pen/Strep. To knock out Kmt2d or Arid1a, the tumor cells were transduced with lentiviruses carrying sgRNAs targeting Kmt2d or Arid1a. The transduced cells were selected under 3–5 μg/ml puromycin, and the knockout of Kmt2d and Arid1a was confirmed by T7E1 assay.
Validating the role of Kmt2d in multiple cancer models
Cas9-expressing MB49 cells, E0771 cells, B16F10 cells, and Lewis lung cancer cells (LLC) were generated by transduction with lentiviruses carrying EFS-Cas9–2A-BlastR-WPRE and selected under 10 μg/ml blasticidin S. To knock out Kmt2d, these Cas9-expressing cells were transduced with lentiviruses carrying an Kmt2d sgRNA, and cells transduced with lentiviral vector or non-targeting sgRNA were used as a control. The transduced cells were selected under 3–5 μg/ml puromycin at 24 h post-infection. To generate syngeneic mouse bladder tumors, 5*106 of vector- or Kmt2d sgRNA- transduced MB49 cells were transplanted subcutaneously into the right flank of C57BL/6J mice. To generate orthotopic breast tumors, 2*106 of vector- or Kmt2d sgRNA- transduced E0771 cells were transplanted into the fatpad of C57BL/6J mice. To generate melanoma or Lewis lung tumors in C57BL/6J mice, 2*106 of vector- or Kmt2d sgRNA- transduced B16F10 cells or Lewis lung cancer cells were subcutaneously transplanted into the right flank of C57BL/6J mice. Tumor growth was monitored and assigned into 2 groups to receive the treatment of anti-PD1 or PBS at the indicated time.
Dual-tumor model of LLC-Vector and LLC-sgKmt2d cells.
To generate a dual-tumor model of LLC, 2*106 of LLC-Vector and LLC-sgKmt2d cells were transplanted into the left flank and right flank of C57BL/6J mice respectively. Tumor growth was monitored, and mice were assigned into 2 groups to receive anti-PD1 or PBS.
Flow cytometry analysis and sorting.
All antibodies for flow were purchased from Biolegend or eBiosciences. Single-cell suspensions of tumors or spleens were prepared using a gentleMACS tissue dissociation system. All flow antibodies were used at 1:100 dilutions for staining unless otherwise noted. After staining, cells were centrifuged at 300–600 g for 5 min, and washed twice with staining buffer before being analyzed or sorted on a BD FACSAria. The data was analyzed using the FlowJo software (v9.9.4 or v10.3). A previously reported strategy was used to define the immune populations in tumor (43).
Mutagenesis with lentiviral CRISPR.
The CRISPR knockout construct Lenti-U6-sgBsmBI-EFS-Puro-WPRE was generated. To clone sgRNA targeting individual genes, such as Kmt2d and Arid1a, the corresponding oligos were synthesized, annealed and cloned into BsmBI linearized lentiviral knockout vectors. The following sgRNAs were used for Kmt2d, sgRNA1: GCCGGCTATGTCGGGCCTGT; sgRNA3: GTGTGTGAGACATGTGACAA. For Arid1a, sgRNA4: GACGCATGAGCCATTCTCCC. Lentiviruses were produced by co-transfecting the lentiviral CRISPR knockout plasmids, together with packing plasmids pMD2.G and psPAX2, into 80–90% confluent HEK293FT cells. The lentivirus-containing supernatants were collected at 48 h and 72 h post-transfection, aliquoted and stored at −80°C before use. To knockout KMT2D in human lung cancer H1299 cells, the corresponding oligos of the sgRNA1: GGTGGAAATTCCCGCCAACG; sgRNA2: AAATGGCTGTTGATCCCATG were synthesized, annealed and cloned into BsmBI linearized lentiviral knockout vectors. After lentiviral production, the Cas9-tranduced tumor cells were infected and selected under 3–5 μg/ml puromycin to obtain individual gene knockout cells. CRISPR mutagenesis was confirmed by extracting genomic DNA for T7E1 assays.
Immunoblot to quantify the levels of ubiquitinated proteins.
Vector, sgKmt2d, or sgArid1a transduced primary liver tumor cells (MA1L), MB49-Vector, MB49-sgKmt2d, LLC-Vector, and LLC-sgKmt2d cells were seeded into 6-well plates or 10-cm dishes and cultured for 24 h. Then, 15 μM MG132, 10ng/ml IFN-γ, or DMSO was added and incubated for 2–3 h before harvesting the treated cells. The harvested cells were washed twice with ice cold PBS, and then lysed with 1× RIPA buffer on ice for 15 min. Cell lysates were centrifuged at 12,000 g for 15 min at 4°C and protein-containing supernatant was collected. Protein concentration was measured using a BCA assay (Abcam) and ~20 μg of proteins from each sample were loaded into SDS-PAGE gel. After electrophoresis and transmembrane, immunoblot assay were performed with antibody against ubiquitin (clone Ubi-1, Sigma), with GAPDH being used as internal control. The relative levels of ubiquitinated protein were quantified by greyscale analysis.
Flow cytometry to quantify cell surface MHC-I and peptide-MHC-I complex.
2*105 of vector, sgKmt2d, or sgArid1a transduced MA1L primary liver tumor cells (MA1L-Vector, MA1L-sgKmt2d, MA1L-sgArid1a), MB49-Vector and MB49-sgKmt2d cells were seeded into 12-well plates. To test the effect of IFN-γ on surface MHCI or peptide-MHCI presentation, 0, 5 ng/ml, or 10 ng/ml IFN-γ were added and treated for 24–48 h. The treated cells were collected and washed twice with 2% FBS in PBS. Then, the cells were stained with 1:100 diluted PE-H-2Kb/H-2Db, and APC-SIINFEKL-H-2Kb for 30 min on ice and washed twice with 2% FBS in PBS before flow cytometry analysis. Samples were run on Attune NxT Flow Cytometer and the mean fluorescence intensity were quantified.
RNA extraction, reverse transcription, and quantitative PCR.
RNA from control and Kmt2d-mutant cells was extracted using TRIzol Reagent (Invitrogen) by following standard RNA extraction protocols. The first-strand cDNA of RNA was synthesized using SuperScript™ IV Reverse Transcriptase (Invitrogen). After normalizing the concentrations of cDNA with nuclease-free water, quantitative PCR (qPCR) was performed by adding designated Taqman probe of interested genes, and GAPDH was used as an internal positive control.
Western Blot.
Cells in 6-well plate or 10-cm dish were washed twice with ice cold PBS. The cells were then lysed with 1× RIPA buffer on ice for 15 min, or nuclear protein purification using nuclear extraction kit (Abcam). Cell lysates were centrifuged at 12,000 g for 15 min at 4°C and protein-containing supernatant was collected. Protein concentration was measured using a BCA assay (Abcam) and 20 μg of protein in each sample were loaded into SDS-PAGE gel. After electrophoresis, proteins separated in gel were transferred into nitrocellulose membranes. Membranes were blocked at room temperature for 1 h using 5% skim-milk in TBST, followed by the incubation with primary antibody in 4°C overnight. After washing three times with TBST, horseradish peroxidase (HRP)-conjugated secondary antibody was added and incubated at room temperature for 30 – 60 min. The chemiluminescent substrate (Clarity Western ECL Substrate, Bio-Rad) was added on top of blot membrane according to the manufacturer’s instructions. The signals were captured using a CCD camera-based imager (GE Healthcare).
MIP sequencing data analysis.
Raw FASTQ reads were mapped to the mm10 genome using bwa mem v.0.7.17 (44). BAM files were sorted and indexed using SAMtools v1.3 (45). Indel variants were then called using SAMtools and VarScan v2.3.9 (46). All detected indels were filtered by requiring that each indel must overlap the ±3 bp window surrounding the predicted cut site of the closest sgRNA. We excluded variants at Rps19 sg5 because vector control samples were also found to have heterozygous mutations at this site.
The remaining indel variants were summed up for each sgRNA site to obtain a mutation frequency table. To further filter detected variants, we employed a false discovery approach based on vector control samples. For each sgRNA in the library, we took the highest variant frequency across all the vector control samples and set this value as the minimum cutoff when filtering the mTSG samples. In addition, we further set a 5% variant frequency cutoff to ensure stringent detection of indels. The filtered sgRNA variant frequency table was then averaged by gene to obtain the gene-level frequency table. We then used the gene-level variant frequencies to determine enrichment or depletion of specific mutations in ICB-treated vs PBS-treated samples by two-tailed unpaired t-test.
Analysis of KMT2D mutation status in patient cohorts.
KMT2D mutation status was queried using cBioPortal using the OQL specifiers “MUT HOMDEL” for all mutations and deletions (47), or “DRIVER NONSENSE NONSTART NONSTOP FRAMESHIFT SPLICE TRUNC HOMDEL” for anticipated loss-of-function mutations and deletions. The different cancer types in the curated non-redundant set were consolidated based on the originating tissue.
To determine the association between KMT2D and tumor mutation burden, the cBioPortal was queried across the PanCancer TCGA cohorts. Tumor types with at least 5 KMT2D-mutant samples were considered for analysis. Statistical significance was assessed by two-tailed Mann-Whitney test.
Analysis of genes and cell types correlated with KMT2D expression in tumors.
RNAseq count data from TCGA were downloaded from the GDC Data Portal and normalized to TPM. The Spearman correlation between each gene and KMT2D was calculated, and p-values were adjusted for multiple comparisons by the Benjamini-Hochberg method. We then tabulated the number of individual cancer types for which a given gene was concordantly correlated with KMT2D. Inferred cell type abundances in TCGA tumors were downloaded from the xCell website (http://xcell.ucsf.edu/). Correlations between KMT2D and cell type abundances were calculated in the same manner as with individual genes.
To obtain the pan-cancer gene sets that are positively or negatively correlated with KMT2D, we selected for genes that are concordantly correlated with KMT2D across multiple cancer types. Based on the empirical cumulative density function of the number of cancer types for which each gene was significantly correlated with KMT2D, we selected a cutoff that would select approximately the top 5% of genes (30+ cancer types among positively correlated genes, 21+ cancer types among negatively correlated genes). DAVID gene ontology analysis was performed on the resultant gene sets.
Exome sequencing analysis.
Raw FASTQ reads were mapped to the mm10 genome using the bwa mem function in BWA v.0.7.17. Mutations in MA1L cells were called using Strelka v2.9.2 by comparing to wildtype liver exomes from C57BL/6J mice.
ATAC-seq analysis.
Raw FASTQ reads were mapped to the mm10 genome using Bowtie v2 (48). ATAC-seq accessible regions were called using MACS2 (49). Accessible regions across all samples were combined and the read counts in each region were tabulated. Pairwise Spearman correlations were calculated using the read counts in each region. Differential accessibility was performed using DESeq2 (50). Intersection of accessible regions and motif analysis was performed using HOMER (51).
RNA-seq analysis.
Raw FASTQ reads were quantified to the mm10 transcriptome using Kallisto (52). Differential expression analysis was performed using Sleuth (53). DAVID gene ontology analysis was performed on genes with an adjusted p < 0.05. For neoantigen prediction, RNA-seq reads were aligned to the mm10 genome using STAR, then mutations were called using the RNA-seq mode of Strelka v2 (54). The resultant mutations were annotated using VEP, then neoantigens were predicted using pVACtools with H-2Kb and H-2Db as the candidate MHCI alleles (55).
To analyze expression of transposable elements, the raw FASTQ reads were first re-aligned using STAR (56) with modified settings (outFilterMultimapNmax 100, winAnchorMultimapNmax 100). Transposable elements were quantified by TEcount from TEToolkit. Differential expression was assessed using the raw counts of all genes and transposable elements with DEseq2.
Analysis of FOSL2 binding in human liver cancer cells.
FOSL2 ChIP-seq data in HepG2 cells were downloaded from the ENCODE database and visualized in IGV.
Sample size determination.
Sample size was determined according to the lab’s prior work or similar studies in the literature.
Randomization and blinding statements.
In animal experiments, mice were randomized by sex, cage and littermates. In vitro experiments were not randomized or blinded. Investigators were blinded in mouse experiments by labeling cages with generic identifiers. In NGS data analysis, investigators were blinded for initial processing of the original data using key-coded metadata.
Standard statistical analysis.
Data between two groups were analyzed using a two-tailed unpaired t-test. Different levels of statistical significance were accessed based on specific p values and type I error cutoffs (0.05, 0.01, 0.001, 0.0001). GraphPad Prism and R were used for analyses.
Code availability.
Codes used for data analysis or generation of the figures related to this study are available on GitHub (https://github.com/rdchow/immunoMIPS/).
Data and resource availability.
All data generated or analyzed during this study are included in this article and its supplementary information files. Specifically, source data and statistics for non-high-throughput experiments such as flow cytometry, qPCR, protein experiments, and other molecular or cellular assays are provided in Supplemental Tables. Processed data for genomic sequencing (e.g. RNA-seq, exome sequencing, ATAC-seq) and other forms of high-throughput experiments are provided as processed quantifications in Supplemental Datasets. Raw sequencing data have been deposited to NIH Sequence Read Archive (SRA) or Gene Expression Omnibus: MIPS sequencing of mouse tissue and exome sequencing of MA1L cells (PRJNA634679); ATAC-seq and RNA-seq (GSE151227). Original cell lines are available at commercial sources listed in supplementary information files. Genetically modified cell lines are available via Chen lab. Most data, reagents, methods, computational codes and materials that support the findings of this research are available from the corresponding author upon reasonable request. Some material used in the reported research may require requests to collaborators and agreements with other entities. Requests are reviewed by Yale University to verify whether the request is subject to any intellectual property or confidentiality obligations. Any material that can be shared will be released via a Material Transfer Agreement.
Supplementary Material
Significance.
Immune checkpoint blockade (ICB) is ineffective in the majority of patients. Through direct in vivo CRISPR mutagenesis screening in GEMMs of cancer, we find Kmt2d deficiency sensitizes tumors to ICB. Considering the prevalence of KMT2D mutations, this finding potentially has broad implications for patient stratification and clinical decision making.
Acknowledgments
We thank Drs. Charles Fuchs and Roy Herbst for discussion. We thank Sarah Slavoff and Zhenkun Na for their assistance in protein work. We thank all members in Chen laboratory, as well as various colleagues in Department of Genetics, Systems Biology Institute, Cancer Systems Biology Center, MCGD Program, Immunobiology Program, BBS Program, Cancer Center, Stem Cell Center, Liver Center, RNA Center and Center for Biomedical Data Sciences at Yale for assistance and/or discussion. We thank the Center for Genome Analysis, Center for Molecular Discovery, Pathology Tissue Services, Histology Services, Electron Microscopy, High Performance Computing Center, West Campus Analytical Chemistry Core and West Campus Imaging Core and Keck Biotechnology Resource Laboratory at Yale, for technical support.
SC is supported by Yale SBI/Genetics Startup Fund, NIH/NCI/NIDA (DP2CA238295, 1R01CA231112, U54CA209992–8697, R33CA225498, RF1DA048811), Damon Runyon Dale Frey Award (DFS-13–15), Melanoma Research Alliance (412806, 16–003524), St-Baldrick’s Foundation (426685), Breast Cancer Alliance, Cancer Research Institute (CLIP), the 2017 AACR NextGen Grant for Transformative Cancer Research (Grant Number 17–20-01-CHEN), The Mary Kay Foundation (017–81), The V Foundation (V2017–022), Ludwig Family Foundation, DoD (W81XWH-17–1-0235 and W81XWH-20–1-0072), Sontag Foundation, and Chenevert Family Foundation. GW is supported by CRI Irvington and RJ Anderson Postdoctoral Fellowships. XD is supported by Charles H. Revson Senior Postdoctoral Fellowship. RDC is supported by the NIH/NCI (T32GM007205, F30CA250249). MBD is supported by the Yale MSTP training grant from NIH (T32GM007205). PR is supported by Yale PhD training grant from NIH (T32GM007499) and Lo Fellowship of Excellence of Stem Cell Research.
Footnotes
Conflict of interest disclosure statement
The authors declare no competing interest related to this work. SC is a co-founder, funding recipient and scientific advisor of EvolveImmune Therapeutics, which is not related to this study.
References
- 1.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. N Engl J Med. 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahmer JR, Tykodi SS, Chow LQM, Hwu W-J, Topalian SL, Hwu P, et al. Safety and Activity of Anti–PD-L1 Antibody in Patients with Advanced Cancer. N Engl J Med. 2012;366:2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, et al. First-Line Nivolumab in Stage IV or Recurrent Non–Small-Cell Lung Cancer. N Engl J Med. 2017;376:2415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powles T, Durán I, van der Heijden MS, Loriot Y, Vogelzang NJ, De Giorgi U, et al. Atezolizumab versus chemotherapy in patients with platinum-treated locally advanced or metastatic urothelial carcinoma (IMvigor211): a multicentre, open-label, phase 3 randomised controlled trial. The Lancet. 2018;391:748–57. [DOI] [PubMed] [Google Scholar]
- 7.Allen EMV, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su M-J, Melms JC, et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell. 2018;175:984–997.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 2018;33:843–852.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miao D, Margolis CA, Vokes NI, Liu D, Taylor-Weiner A, Wankowicz SM, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet. 2018;50:1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harel M, Ortenberg R, Varanasi SK, Mangalhara KC, Mardamshina M, Markovits E, et al. Proteomics of Melanoma Response to Immunotherapy Reveals Mitochondrial Dependence. Cell. 2019;179:236–250.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell. 2018;175:998–1013.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. 2018;359:770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547:413–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samstein RM, Lee C-H, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. [DOI] [PubMed] [Google Scholar]
- 18.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen P-L, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov. 2016;6:827–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer. 2018;18:139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer. 2018;118:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348:74–80. [DOI] [PubMed] [Google Scholar]
- 24.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:654–8. [DOI] [PubMed] [Google Scholar]
- 26.Chow RD, Guzman CD, Wang G, Schmidt F, Youngblood MW, Ye L, et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci. 2017;20:1329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Chow RD, Ye L, Guzman CD, Dai X, Dong MB, et al. Mapping a functional cancer genome atlas of tumor suppressors in mouse liver using AAV-CRISPR–mediated direct in vivo screening. Sci Adv. 2018;4:eaao5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kantidakis T, Saponaro M, Mitter R, Horswell S, Kranz A, Boeing S, et al. Mutation of cancer driver MLL2 results in transcription stress and genome instability. Genes Dev. 2016;30:408–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon SR, Maute RL, Dulken BW, Hutter G, George BM, McCracken MN, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saponaro M, Kantidakis T, Mitter R, Kelly GP, Heron M, Williams H, et al. RECQL5 Controls Transcript Elongation and Suppresses Genome Instability Associated with Transcription Stress. Cell. 2014;157:1037–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge Z, et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat Med. 2018;24:556–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snyder A, Nathanson T, Funt SA, Ahuja A, Buros Novik J, Hellmann MD, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: An exploratory multi-omic analysis. PLoS Med. 2017;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laumont CM, Daouda T, Laverdure J-P, Bonneil É, Caron-Lizotte O, Hardy M- P, et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat Commun. 2016;7:10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laumont CM, Vincent K, Hesnard L, Audemard É, Bonneil É, Laverdure J-P, et al. Noncoding regions are the main source of targetable tumor-specific antigens. Sci Transl Med. 2018;10:eaau5516. [DOI] [PubMed] [Google Scholar]
- 38.Smart AC, Margolis CA, Pimentel H, He MX, Miao D, Adeegbe D, et al. Intron retention is a source of neoepitopes in cancer. Nat Biotechnol. 2018;36:1056–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yewdell JW, Antón LC, Bennink JR. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J Immunol. 1996;157:1823–6. [PubMed] [Google Scholar]
- 40.Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee J-E, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strauss L, Mahmoud MAA, Weaver JD, Tijaro-Ovalle NM, Christofides A, Wang Q, et al. Targeted deletion of PD-1 in myeloid cells induces antitumor immunity. Sci Immunol. 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varghese F, Bukhari AB, Malhotra R, De A. IHC Profiler: An Open Source Plugin for the Quantitative Evaluation and Automated Scoring of Immunohistochemistry Images of Human Tissue Samples. PLOS ONE. 2014;9:e96801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broz ML, Binnewies M, Boldajipour B, Nelson AE, Pollack JL, Erle DJ, et al. Dissecting the Tumor Myeloid Compartment Reveals Rare Activating Antigen-Presenting Cells Critical for T Cell Immunity. Cancer Cell. 2014;26:638–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 53.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14:687–90. [DOI] [PubMed] [Google Scholar]
- 54.Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Källberg M, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15:591–4. [DOI] [PubMed] [Google Scholar]
- 55.Hundal J, Kiwala S, McMichael J, Miller CA, Xia H, Wollam AT, et al. pVACtools: a computational toolkit to identify and visualize cancer neoantigens. Cancer Immunol Res. 2020; 8:409–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.