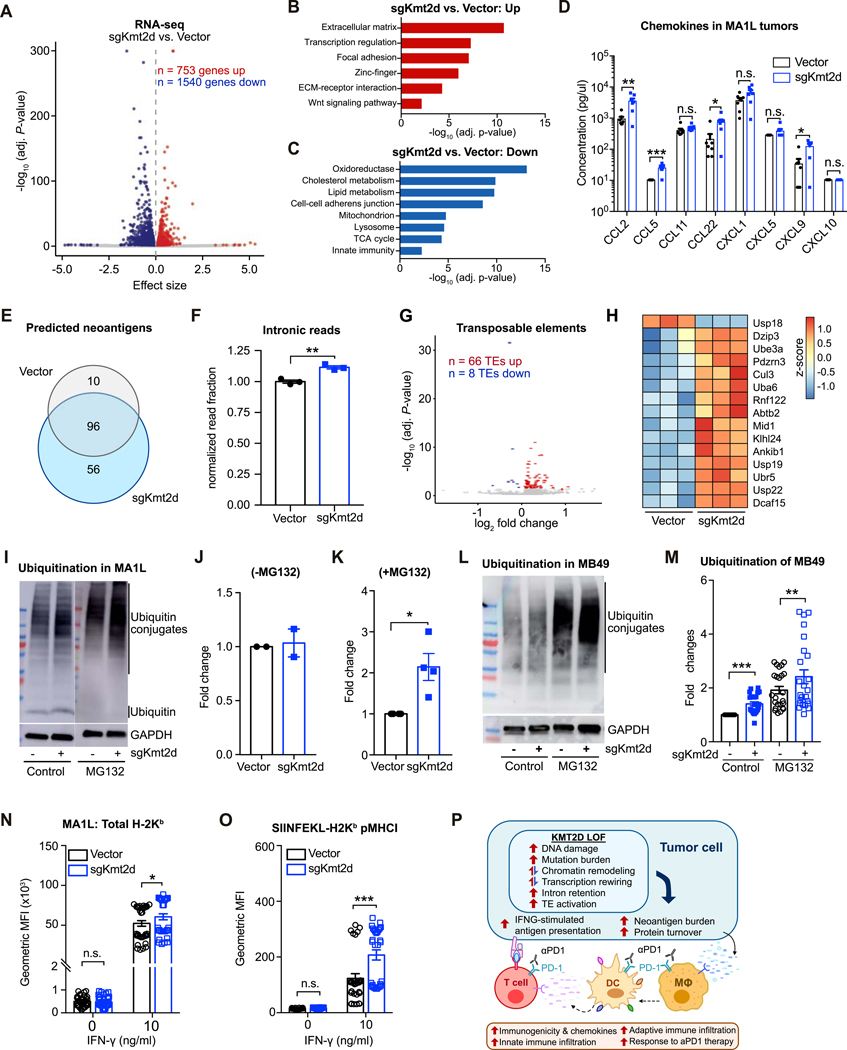
Figure 7. Pleiotropic effects of Kmt2d deficiency on transcriptional regulation, protein turnover, and antigen presentation.
(A) Volcano plot of RNA-seq data, comparing MA1L cells transduced with sgKmt2d (n = 3) vs. Vector (n = 3). 753 genes were significantly upregulated, while 1540 genes were downregulated (adjusted P < 0.05).
(B-C) DAVID gene ontology analysis of genes significantly upregulated (B) or downregulated (C) with Kmt2d deficiency.
(D) Chemokines in the tumors formed by MA1L-Vector and MA1L-sgKmt2d were profiled using the LEGENDplex™ Mouse Proinflammatory Chemokine Panel (13-plex). T-test between MA1L-sgKmt2d (n = 7) vs. MA1L-Vector (n = 7): CCL2, P = 0.0072; CCL5, P = 0.0003; CCL11, P = 0.1961; CCL22, P = 0.0202; CXCL1, P = 0.0948; CXCL5, P = 0.1322; CXCL9, P = 0.0105.
(E) Venn diagram of predicted neoantigens in Vector vs. sgKmt2d transduced cells.
(F) Relative proportion of RNA-seq reads mapping to intronic regions, comparing Vector (n = 3) vs. sgKmt2d (n = 3) transduced cells. Two-tailed unpaired t-test, P = 0.0033.
(G) Volcano plot of transposable element (TE) expression profiles by RNA-seq, comparing MA1L cells transduced with sgKmt2d (n = 3) vs. Vector (n = 3). 66 TEs were significantly upregulated, while 8 TEs were downregulated (adjusted P < 0.05).
(H) Heat map of top differentially expressed genes involved in protein turnover, shown as z-scores.
(I) Western blot analysis of ubiquitin (conjugated or free form) and GAPDH in Vector or sgKmt2d-transduced MA1L liver cancer cells, with (+MG132, right panel) or without (-MG132, left panel) proteasome inhibitor treatment.
(J) Quantification of ubiquitin conjugates in the absence of proteasome inhibition, normalized to Vector.
(K) Quantification of ubiquitin conjugates in the presence of proteasome inhibition, normalized to Vector. Paired t-test, sgKmt2d (n = 4) vs. Vector (n = 4), P = 0.0403.
(L) Western blot analysis of ubiquitin conjugates and GAPDH in Vector or sgKmt2d transduced MB49 bladder cancer cells, with or without the addition of proteasome inhibitor MG132.
(M) Quantification of ubiquitin conjugates in Kmt2d-mutant MB49 bladder cancer cells with or without MG132, normalized to the vector control. Two-tailed paired t-test, without MG132: sgKmt2d (n = 24) vs. Vector (n = 24), P < 0.0001; with MG132: sgKmt2d (n = 24) vs. Vector (n = 24), P = 0.0054.
(N) Flow cytometry analysis of total H-2Kb expression levels in Vector vs. sgKmt2d transduced cells, cultured in 0 ng/ml or 10 ng/ml IFN-γ. Two-tailed unpaired t-test, Vector (n = 36) vs. sgKmt2d (n = 36): 0 ng/ml IFN-γ, P = 0.9931; 10 ng/ml IFN-γ, P = 0.0250.
(O) Flow cytometry analysis of SIINFEKL-H-2Kb peptide-MHC-I complexes in Vector vs. sgKmt2d transduced cells, cultured in 0 ng/ml or 10 ng/ml IFN-γ. Two-tailed unpaired t-test, Vector (n = 30) vs. sgKmt2d (n = 30): 0 ng/ml IFN-γ, P = 0.9109; 10 ng/ml IFN-γ, P < 0.0001. Data of (N-O) were collected from 4 independent experiments
(P) Schematic summarizing the pleiotropic consequences of Kmt2d deficiency on tumor cell-intrinsic properties, leading to increased immune infiltration and potentiating response to aPD1 immunotherapy.
Error bars: All data points in this figure are presented as mean ± SEM. Asterisks: * P < 0.05, ** P < 0.01, *** P < 0.001.
See also: Supplementary Fig. S10 and S11