Figure 1. Tissue-Specific Metabolic Adaptations of T Cells.
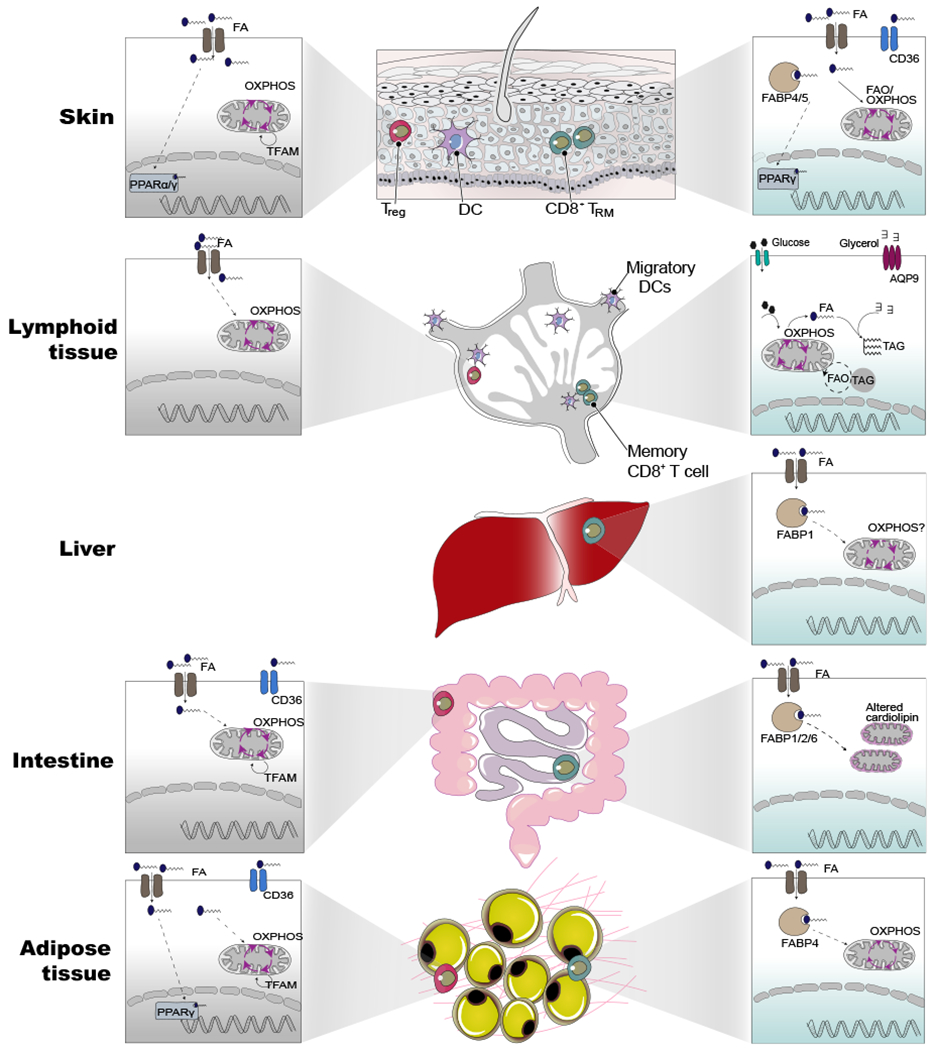
Tissue-resident memory and Tregs from different tissues share a core metabolic program involving in fatty acid metabolism and mitochondrial metabolism compared with their counterparts in lymphoid tissues. Beyond this core metabolic program, each population possesses additional tissue-specific programs shaped by the local environment. For instance, skin TRM show the expression of PPARγ and express fatty-acid-binding protein isoform, FABP4/5, whereas liver TRM and adipose tissue express the isoform FABP1 and FABP4, respectively. Another notable difference is the altered cardiolipin content of mitochondrial membranes in the intestinal interepithelial lymphocytes. Interestingly, skin Treg cells express either PPARα/γ isoforms whereas, adipose-tissue-specific Tregs express only PPARγ. However, Treg cells in non-lymphoid tissues, but not in the lymphoid tissues, depended on mitochondrial DNA replication and transcription orchestrated by Tfam. For each tissue, we have shown Treg (gray box) and CD8+ TRM (green box) depicting known metabolic pathways or genes related to metabolism. The dotted lines indicate mechanism/pathway that are unclear.
