Figure 4. Mapping of molecular domains/motifs and structure-based modeling and simulations reveal critical regions and interfacial interactions involved in the complex formation between B7-H4 and E3 ligase AMFR.
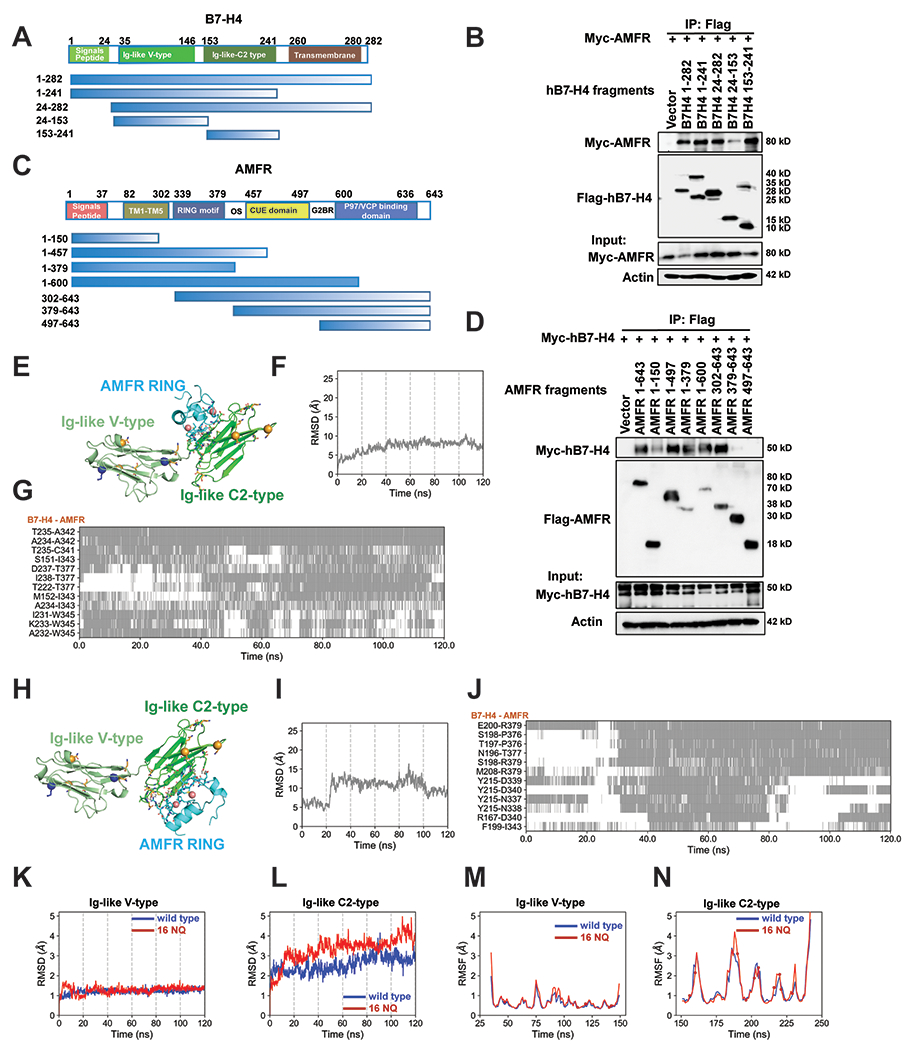
(A) Schematic diagram of human B7-H4 domains and strategy to engineer a series of B7-H4 deletion mutants. (B) Mapping of B7-H4 regions (sequence ranges) involved in interactions with AMFR. The interactions between Myc-AMFR and the displayed Flag-hB7-H4 fragments were examined by co-IP experiments in 293T cells. (C) Schematic diagram of human AMFR domains/motifs and strategy to engineer a series of AMFR deletion mutants. Abbreviations: TM (transmembrane domain), RING motif (E3 ligase activity), OS (Oligomerization domain), CUE domain (Couples Ubiquitin molecules to ER degradation), G2BR (Ube2G2 binding region), VIM (p97/VCP interacting motif). (D) Mapping of AMFR domains/motifs that interact with B7-H4. The interactions between Myc-hB7-H4 and the displayed Flag-AMFR fragments were examined by co-IP experiments in 293T cells. The representative structures of the top 1 cluster of the ZDOCK docking poses between B7-H4 and AMFR RING domain are shown in (E). AMFR RING, B7-H4 Ig-like V type and Ig-like C2-type domains are in cyan, pale green and green. Ubiquitination sites (K138 and K146) are shown as blue sticks with alpha-carbon atoms highlighted in blue spheres. All asparagines are shown as orange sticks, and the alpha-carbon atoms of five identified asparagines N112, N140, N156, N160, N255 (not shown in the structure) are highlighted in orange spheres. The two ZN2+ ions form coordination bonds are shown as salmon spheres. Residues in the binding interfaces from AMFR and B7-H4 are shown as cyan and green sticks, respectively. Time evolution of the RMSDs of AMFR RING domain in the 120 ns MD simulations of the complex formed with B7-H4 using the start points in (E) is shown in (F). The RMSD was evaluated after structurally aligning the conformers observed during MD trajectories with respect to the B7-H4 Ig-like C2-type domain. The corresponding time evolution of residue-residue interactions between B7-H4 and AMFR RING domain residues are shown in (G). Regions shaded in gray refer to time intervals during which the indicated atom pairs (ordinate) made interfacial contacts. The representative structures of the top 2 cluster of the ZDOCK docking poses between B7-H4 and AMFR RING domain are shown in (H). Time evolution of the RMSDs of AMFR RING domain in the 120 ns MD simulations of the complex formed with B7-H4 using the start points in (H) is shown in (I). The corresponding time evolution of residue-residue interactions between B7-H4 and AMFR RING domain residues are shown in (J).The RMSDs profiles of the B7-H4 Ig-like (K) V-type and (L) C2-type domains are shown in blue and red curves for B7-H4 wildtype and 16NQ mutant, respectively. The corresponding RMSFs values of residues in the two domains are shown in (M) and (N), respectively. The spheres on the red curves indicate the positions of the 16NQ mutations. The RMSDs and RMSFs values for the 16NQ mutant are slightly higher than those of the wildtype in the C2-type domain, though the difference is not statistically significant. Flexibilities (RMSDs and RMSFs) of the Ig-like V-type domain of B7-H4 wildtype and16NQ mutant are quite similar.
