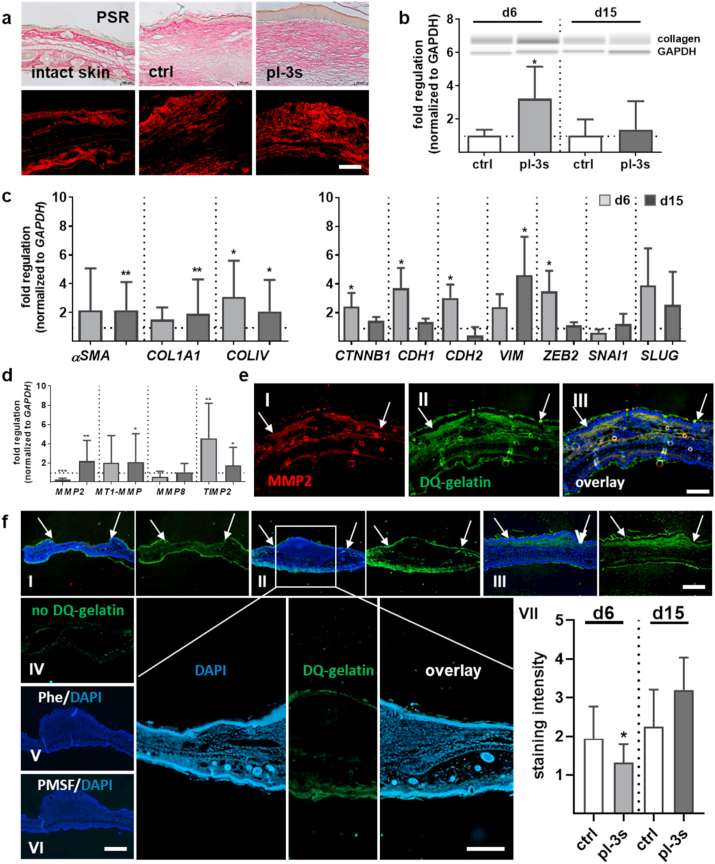
Fig. 4.
Plasma-driven cellular transformation and assessment of gelatinolytic activity during wound healing. (a) PSR staining of collagen fibers in gas plasma-treated wound tissue compared to untreated control showing granulation tissue with well-organized collagen fibers in parallel bundles. (b) Quantification of collagen expression levels as determined by Western blot analysis. (c) αSMA, COL1A1, and COLV gene expression levels were determined after 6 and 15 d following gas plasma treatment using qPCR and corresponding primers. Expression of characteristic EMT markers including β1-catenin (CTNNB1), epithelial E-cadherin (CDH1), mesenchymal N-cadherin (CDH2), vimentin (VIM), melanocytic transcription factor zinc finger E-box-binding homeobox 2 (ZEB2), and the transcription factors SNAI1 and SLUG after 6 d and 15 d following gas plasma treatment using qPCR and corresponding primers. (d) qPCR of MMP2, MT1-MMP, and their inhibitor TIMP2 using qPCR. (e) Co-localization of MMP2 activity (red, I) and gelatinase (green, II) using immunostaining with anti-MMP2 primary and Alexa 594 secondary antibodies (III, merge). (f) The gelatinolytic activity of MMP2/9 was determined using in situ zymography utilizing DQ gelatin (green) in PFA-sections. Representative images of gelatinolytic activity on d0 (I), d6 (II, and higher magnification with scale bar = 200 μm), and d15 (III). In situ zymography without DQ gelatin (green autofluorescence, IV), with a metalloproteinase inhibitor 1,10-phenanthroline (Phe, V), and a serine protease inhibitor PMSF (VII). The gelatinolytic activity of gelatinases was quantified in controls vs. gas plasma-treated samples on d6 and d15 by scoring of the signaling intensity between 1 and 10 (VII). The cell nuclei were counterstained with DAPI (blue), and the wound area was marked with arrows. Scale bars are 50 μm (a), 100 μm (e). Data were normalized to GAPDH and untreated controls (ctrl) and presented as mean +SE. Statistical analysis was done by unpaired, two-tailed Student's t-test (n > 4) with *p < 0.05, **p < 0.01, and ***p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)