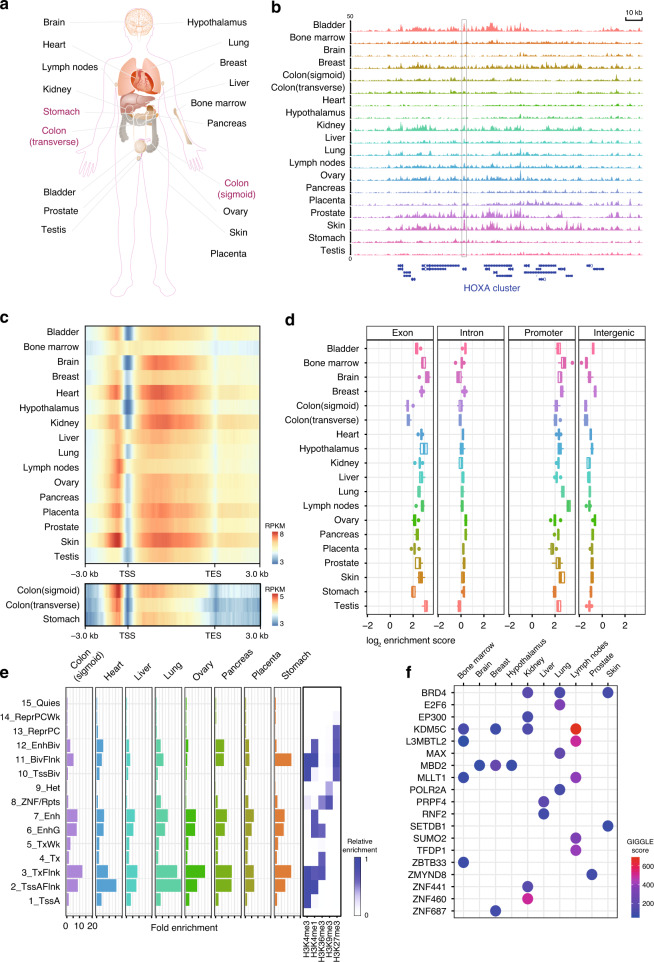
Fig. 1. Landscape of 5hmC across different tissues.
a Schematic plot showing all the organ tissues analyzed in this study. Tumor adjacent tissues are marked in red. b Genome Browser view depicting 5hmC genomic distributions at the HOXA gene cluster for one representative donor’s profile for each of the 19 tissue types assayed. The peak outlined with a box shows a highly variable region as an example. c Metagene plot of 5hmC profiles across different tissues showing underrepresentation of 5hmC at TSS regions and over-representation at gene bodies and promoters. The color ranges indicate RPKM values. Upper panel, normal tissues; lower panel, tumor adjacent tissues. TSS, transcription start site. TES, transcription end site. d Boxplots showing genomic enrichment of 5hmC peaks at RefSeq exons, introns, promoters, and intergenic regions. N = 5 biologically independent samples were used (n = 4 for hypothalamus and n = 6 for sigmoid and transverse colon). For all boxplots, center line represents median, bounds of box represent 25th and 75th percentiles and whiskers are Tukey whiskers. e Enrichment of 5hmC peaks on 15 chromHMM chromatin states from the Roadmap Epigenomics Project for 8 tissue types. Histone modification emissions data are directly from the Roadmap Epigenomics Project. f Enrichment of trans-acting factors binding sites on 5hmC peaks in different tissues. Higher GIGGLE score means higher possibility of enrichment.