Summary
Conjugative pili are important in mediating bacterial conjugation and horizontal gene transfer. Since plasmid transfer can include antibiotic resistance genes, conjugation is an important mechanism in the spread of antibiotic resistance. Filamentous bacteriophages have been shown to exist in two different structural classes: those with a 5-fold rotational symmetry, and those with a 1-start helix with ~ five subunits per turn. Structures for the F and the F-like pED208 conjugation pilus have shown that they have 5-fold rotational symmetry. Here, we report the cryo-EM structure of conjugative pili from carbapenem-resistant Klebsiella pneumoniae, encoded on the IncFIIK pKpQIL plasmid, at 3.9 Å resolution and show that it has a 1-start helix. These results establish that conjugation pili can exist in at least two structural classes, consistent with other results showing that relatively small perturbations are needed to change the helical symmetry of polymers.
Graphical Abstract
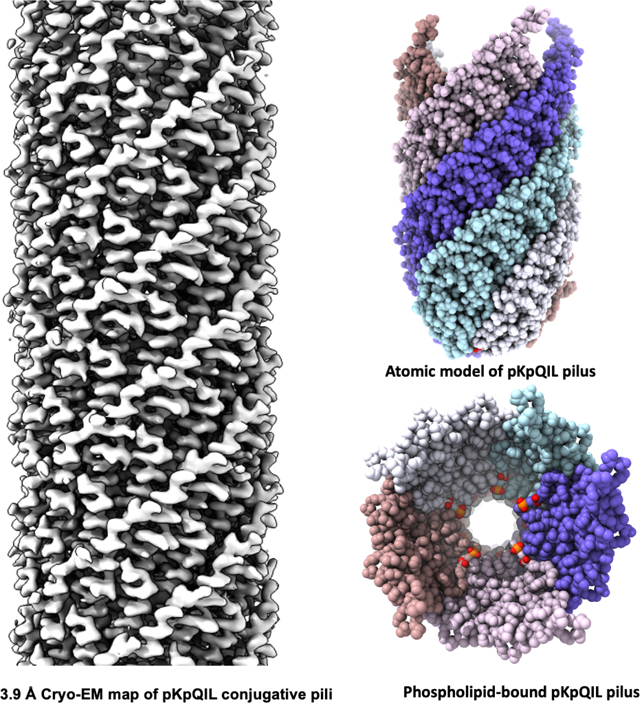
eTOC
Filamentous bacteriophage have been known for many years to exist in two structural classes, those with 5-fold rotational symmetry and those with no rotational symmetry. Zheng et al. show that mating pili can also exist in these same two structural classes.
Introduction
Conjugation, a process mediating contact-dependent horizontal gene transfer between bacterial cells, is an important means by which antibiotic resistance genes transfer between bacterial pathogens (Waksman, 2019). This process requires a type IV secretion system (T4SS), which includes the conjugation pilus (also called the sex pilus or the mating pilus), the translocation channel protein and an ATPase (Chandran Darbari and Waksman, 2015). The conjugation pilus is of central importance to mediating conjugation. The best characterized conjugative pili, the F-type, have been extensively studied using many different techniques (Anthony et al., 1999; Anthony et al., 1994; Arutyunov and Frost, 2013; Frost et al., 1994; Harris and Silverman, 2004; Malmborg et al., 1997; Manchak et al., 2002),(Clarke et al., 2008; Daehnel et al., 2005; Grossman et al., 1990; Paiva and Silverman, 1996; Silverman, 1997). They are long appendages with an outer diameter of ~85 Å and a ~25 Å diameter lumen, but there’s no direct evidence that ssDNA is transferred through the F-pilus during conjugation. Studies have suggested that the role of the F-pilus is to make the initial contact with another cell and then to depolymerize, thus bringing two cells into physical juxtaposition. In this model, DNA transfer only occurs when the pilus is completely depolymerized (Panicker and Minkley, 1985). A more recent study has shown that DNA transfer can occur when two cells are spatially separated, presumably through the mating pilus (Babic et al., 2008). However, the rate of such long-distance transfers is quite low, suggesting that the main mode of DNA transfer occurs when cells are in physical contact. Since, to date, only F-pili have been shown to actively contract, there are still many questions that remain about the role of other conjugation pili that may not be contractile.
A number of similarities have been noted between conjugation pili and filamentous ssDNA bacteriophage: 1) both are polymerized from relatively small (~ 50–70 residue) proteins that are integral membrane proteins in the inner bacterial membrane prior to polymerization; 2) these proteins are largely α-helical; and 3) the filaments are all ~ 80 Å in diameter with a hollow lumen. Filamentous bacteriophages have been grouped into two classes based upon their structures (Xu et al., 2019). One group, including fd, M13, Ike and If1, has been called class I, and all have 5-fold rotational symmetry, with rings of pentamers stacked on top of each other in a helical manner. The other group, including Pf1, Pf3, and Xf, has been called class II, and these have no rotational symmetry, but rather a 1-start helix with ~ 5.4 subunits per turn. Recently, cryo-EM analysis of the F-like pED208 pilus and the F pOX38 pilus has shown that they have 5-fold rotational symmetry, similar to the class I bacteriophage (Costa et al., 2016). Five pED208 pilins form a planar ring, which is related to the pentamer above by a rotation of 28.2° and rise of 12.1 Å. The N- and C-termini of pED208 TraA pilin are exposed on the outside of the filament. Importantly, the structure also revealed that pili are composed of both pilin and phospholipid in a stoichiometric ratio of 1:1. The phosphate head group of the lipids is exposed in the lumen, making the surface inside the pilus moderately negative. Such an electronegative surface would repel DNA, allowing DNA to flow through the internal channel with minimal resistance (Costa et al., 2016). A similar electronegative lining was seen in the lumen of the bacteriophage T4 contractile tail used to inject dsDNA into cells, while a homologous pyocin structure employed to transport a broad range of ions is rather neutral in comparison (Zheng et al., 2017).
Carbapenem-resistant Klebsiella pneumoniae has emerged as a major class of pathogen that spreads rapidly in health care systems, threatening global public health (Chen et al., 2014a; Lee et al., 2016). The spread of these bacteria is accelerated by antibiotic resistance genes carried on plasmids that can easily be transferred to other strains and species. In particular, the globally pervasive carbapenem-resistant K. pneumoniae sequence type ST258 is largely associated with the blaKPC-encoding IncFIIK pKpQIL-family plasmids (Chen et al., 2014b). We have therefore looked at the conjugative pili encoded by pKpQIL (pKpQIL pili). We show a cryo-EM structure of pKpQIL pili at 3.9 Å resolution with a 1-start helical symmetry, distinct from the 5-fold symmetry of pED208. As with the two classes of symmetry found for filamentous bacteriophage, we have now shown that conjugative pili can also be found in these two classes. These results are consistent with prior structural studies showing that relatively small changes in sequence can be amplified by larger changes in quaternary structure (Egelman et al., 2015).
Results
Cryo-EM reconstruction of pKpQIL pili
We purified the pKpQIL pili form a K. pneumoniae strain harboring a plasmid containing a finO mutation (pKpQILΔfinO), which constitutively expresses the tra operon and the pili (Low et al., 2020). We then solved the structure of pKpQIL pili using cryo-EM (Figure 1A). The three-dimensional reconstruction of pKpQIL pili has a helical rise of 2.7 Å and twist of 77.6° per subunit (Figure 1B). Averaged power spectra of pKpQIL pili (Figure S1A–S1C) and pED208 pili (Costa et al., 2016) are similar in the overall distribution of the layer-lines, highlighting the potential ambiguities in determining helical symmetry from such power spectra (Egelman, 2014). Strikingly, imposing the helical parameters for 5-fold rotational symmetry (Figure S1E) or 1-start helical symmetry (Figure S1D) generated two reasonable maps at ~ 5 Å resolution with clear α-helices. However, only imposing the correct 1-start helical symmetry, with ~ 4.6 subunits per turn, resolved more molecular details such as side chains and the phospholipid, which indicates that for some structures even 5 Å resolution is inadequate to determine the correct helical symmetry (Egelman, 2014). Using the correct helical symmetry a resolution of 3.9 Å was obtained (Fig. S2). The ambiguity in determining the correct helical symmetry from the overall power spectrum (Figure S1A) arises from the unknown amount of out-of-plane tilt. Unfortunately, there is no means of determining the degree of out-of-plane tilt of a helical segment from a projection image without having a model to use as a reference. This creates a circularity: the model that is being used as a reference may have the wrong symmetry, and thus the out-of-plane tilt calculated for every segment may be wrong. The only solution to this problem is trying every possible symmetry until one yields an interpretable high-resolution map. Knowing the correct symmetry for the pKpQIL filaments, one can create two different averaged power spectra: one where all segments with large out-of-plane tilt have been excluded (Figure S1B) and one from only those segments with an out-of-plane tilt equal to or greater than 12° (Figure S1C). The difference between these two power spectra is that when large out-of-plane tilt segments are excluded (Figure S1B) clearly separated peaks on the layer line at 1/(12.5 Å) are seen, indicative of the 1-start helical symmetry. In contrast, the full average (Figure S1A) or the power spectrum from only those segments with large out-of-plane tilt (Figure S1C) have intensity on this layer line at 1/(12.5 Å) that appears meridional, which would arise from an n=0 Bessel function in the case of 5-fold rotational symmetry.
Figure 1. Cryo-EM reconstruction of pKpQIL pili at 3.9 Å.
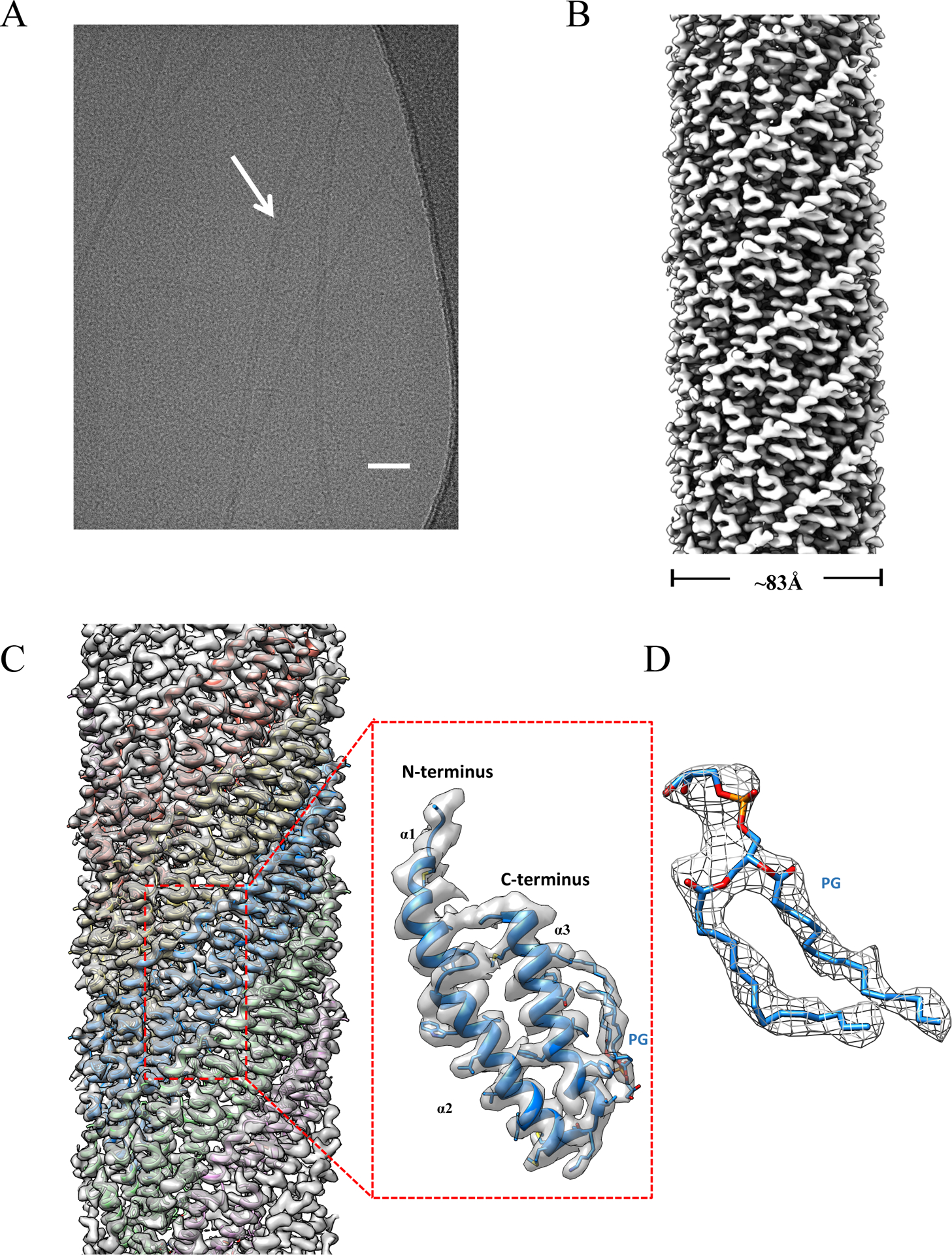
(A) A representative cryo-EM micrograph of pKpQIL pili, scale bar 50nm. (B) Side view of the pKpQIL pili cryo-EM map colored in grey. (C) The cryo-EM map of pKpQIL pili was fit with the atomic model (left), and the five strands are colored in plum, green, blue, yellow and salmon, respectively. A single pilin subunit (right) is shown in ribbon representation (blue), which is composed of three α-helices: α1, α2 and α3. The phospholipid is shown in a stick representation. (D) The density for the phospholipid is shown in mesh.
Morphologically similar to the previously studied pED208 pilus, the pKpQIL pilus is a hollow tube with outer diameter of ~ 83 Å and with a ~ 25 Å diameter inner lumen. The N- and C- termini of the subunits are on the exterior of the pili, while the central portion of the sequence faces into the lumen. The pilin contains 69 amino acids, which result from a cleavage of a signal peptide containing 53 amino acids in the pre-pilin. The first three residues in the pKpQIL pilin N-terminus was disordered and had no density in the cryo-EM map. As with the pED208 pilus, a disconnected density was observed at the subunit-subunit interface that could not be explained by the atomic model for the pilin. This density can be clearly fit as a phospholipid (Figure 1C and 1D). We have not been able to determine the exact lipid present, but have successfully modeled it at the present resolution as phosphatidylglycerol. The pilin subunit consists of three α-helices, designated α1, α2 and α3. It is α3 which mainly binds the phospholipid, largely through interactions of hydrophobic sidechains with the acyl chains of the phospholipid (Figure 1C). But the acyl chains will also interact with α2 in a neighboring subunit (Figure 2G).
Figure 2. Intermolecular interactions within pKpQIL pili.
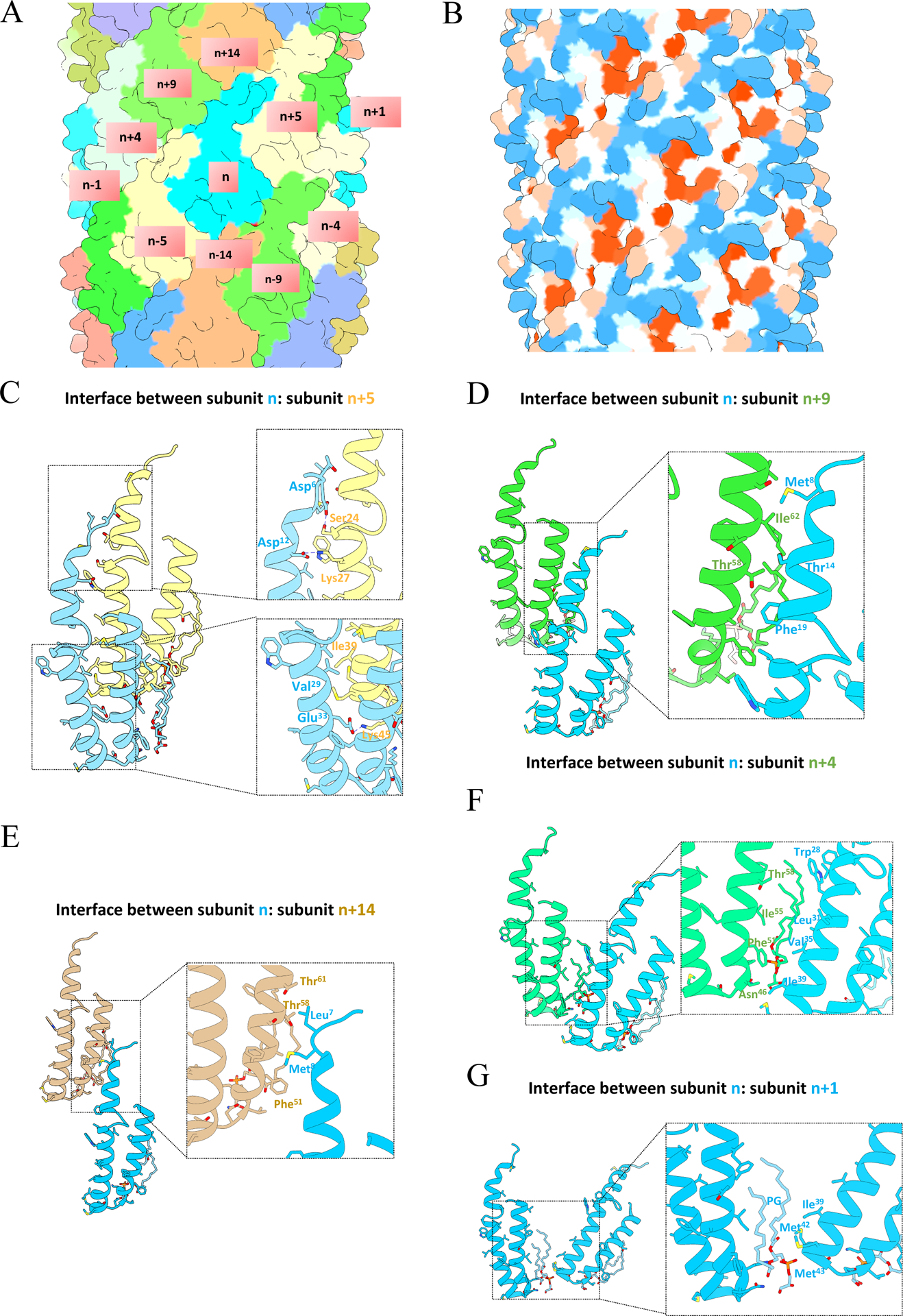
(A) Surface representation of atomic model colored by chains. A subunit n interacts with ten adjacent subunits which are labeled. (B) Hydrophobicity patterns of pKpQIL pili. Hydrophobic residues are shown in red and hydrophilic residues are shown in blue on the surface of the pilus. (C) The interface between subunit n and n+5. (D) The interface between subunit n and n+9. (E) The interface between subunit n and n+4. (F) The interface between subunit n and n+14. (G) The interface between subunit n and n+1.
Structural characterization of intermolecular interactions within pKpQIL pili
The pilin subunits are held together by both hydrophobic and hydrophilic interactions between the neighboring subunits. Labeling each subunit along the 1-start helix, subunit n interacts with ten neighboring subunits: subunit n−1, n+1, n−4, n+4, n−5, n+5, n−9, n+9, n−14, n+14, forming lateral and longitudinal contacts (Figure 2A and 2B). Five different intermolecular interfaces are formed (Figure 2C–2G). Most of the interactions involve hydrophobic interactions (Figure 2D–2G), however, putative hydrogen bonding network are suggested by the atomic model between subunit n and subunit n+5 (Figure 2D). This occurs between the N-terminus of subunit n and subunit n+5, where the Asp 12 of subunit n would form a salt bridge with Lys 27 of subunit n+5 and Asp 6 of subunit n would form a hydrogen bond with Ser 24 of subunit n+5. Also, the interface between subunit n and subunit n+5 has the largest interfacial area among all the different intermolecular interfaces, with a buried surface area of 1,085 Å (Table S1). Therefore, the interaction between subunit n and subunit n+5 appears essential in stabilizing the helical assembly of pKpQIL pili.
Structural comparison of pKpQIL and pED208 conjugative pili
We compare the structure of the pKpQIL pilus with the previously determined structure of the pED208 pilus (Costa et al., 2016). The overall sequence identity between the two conjugative pili is 40% Fig. 3A,B), and not surprisingly, the overall folds of pED208 and pKpQIL pilin are very similar (Figure 3C,D). Structural similarity can be measured by the Dali server (Holm and Rosenstrom, 2010), and the Z-score of 8.6 between the two pilin subunits indicates the high degree of similarity between the two structures. Superposition of the Cα atoms of pED208 pilin and pKpQIL pilin yielded an overall RMSD between the 62 paired residues of only 1.4 Å, which also indicates the high degree of structural similarity (Figure 3C). However, some differences were observed in the monomeric structures as well as the intermolecular interfaces in the pili. The pED208 pilin has an extended loop in the N-terminus, while the N-terminus of the pKpQIL pilin has an extended α-helical structure. As the N-terminus of conjugative pili has been shown to be responsible for phage recognition (Meng et al., 2019), this suggests the two forms of conjugative pili might be selectively recognized by different phages.
Figure 3. Structural comparison of two forms of conjugative pili.
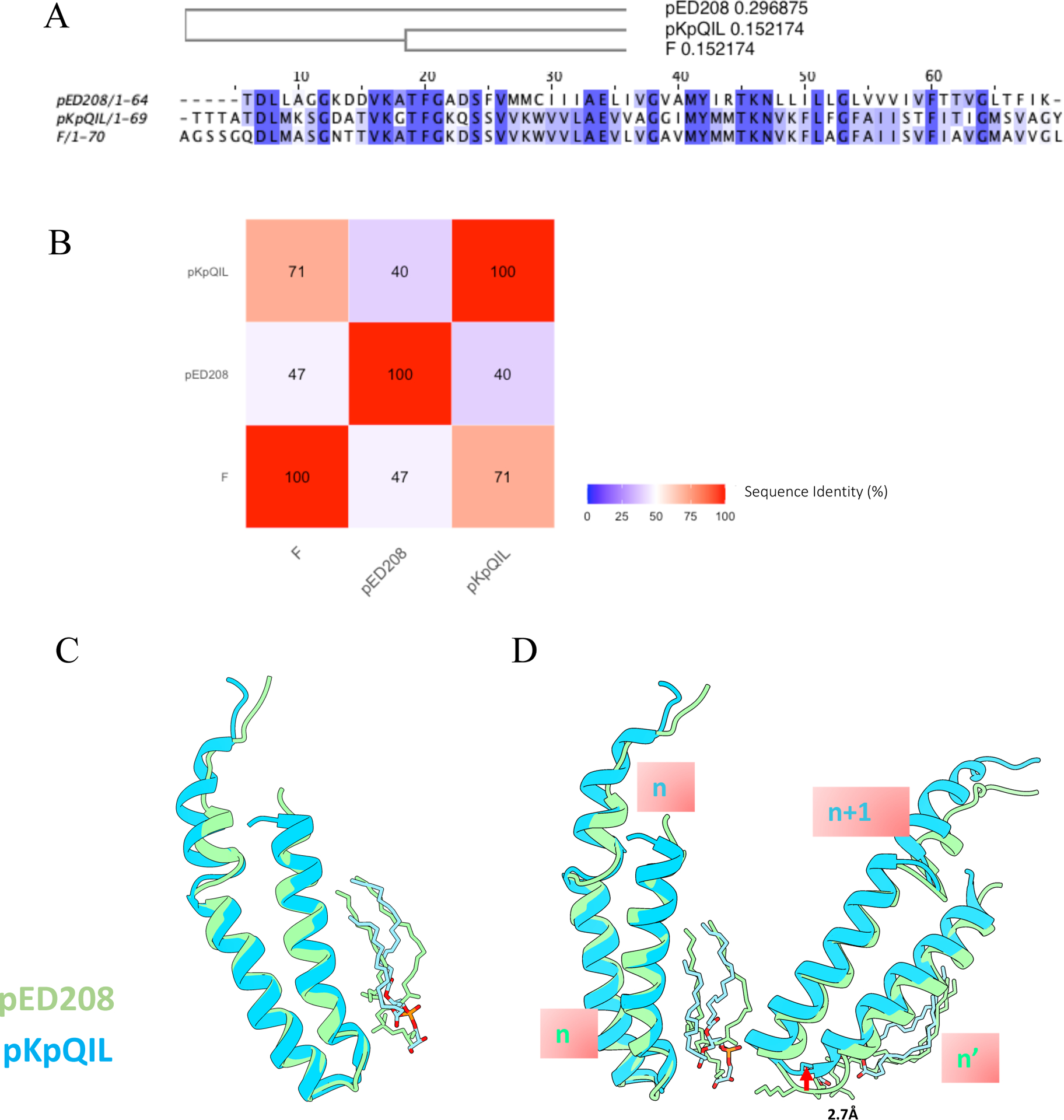
(A) Sequence alignment of pKpQIL, F and pED208 pilins with the phylogenetic tree showing that the sequence of pKpQIL pili is more closely related to F pili than to pED208. (B) Heatmap of sequence identity of pKpQIL, F and pED208 pili. (C) Superposition of pKpQIL pilin subunit and pED208 pilin subunit. (D) Comparison of the packing of adjacent pilin subunits in pKpQIL pili and pED208 pili. When superimposing subunit n from pKpQIL pili and pED208 pili, subunit n+1 in pKpQIL is shifted 2.7 Å along the helical axis relative to the corresponding subunit in pED208 pili.
To further compare the two forms of conjugative pili with distinct symmetries, two adjacent subunits (n and n+1) from pKpQIL pili were compared with two adjacent subunits in the same pentamer layer from pED208 pili (PDB ID: 5LEG). When the first two subunits are aligned, subunit n+1 from pKpQIL is shifted along the helical axis by 2.7 Å compared to the second subunit from pED208 pili (Figure 3D). This minor difference in the packing of adjacent pilin subunits accumulates as subsequent subunits are added, resulting in the completely distinct helical symmetry between the two forms of conjugative pili.
Both pKpQIL pili and pED208 pili are composed of pilin:phospholipid in a 1:1 ratio, and the negatively charged head group of the phospholipids points into the lumen of the pilus (Figure 4A). In both structures, the phospholipids reverse the electrostatic potential of the lumen from positively charged to negatively charged (Figure 4B), which, in any model involving ssDNA transfer through the pilus, would serve to repel DNA from the walls of the lumen.
Figure 4. Phospholipid changes electrostatic surface potential of the pKpQIL pili.
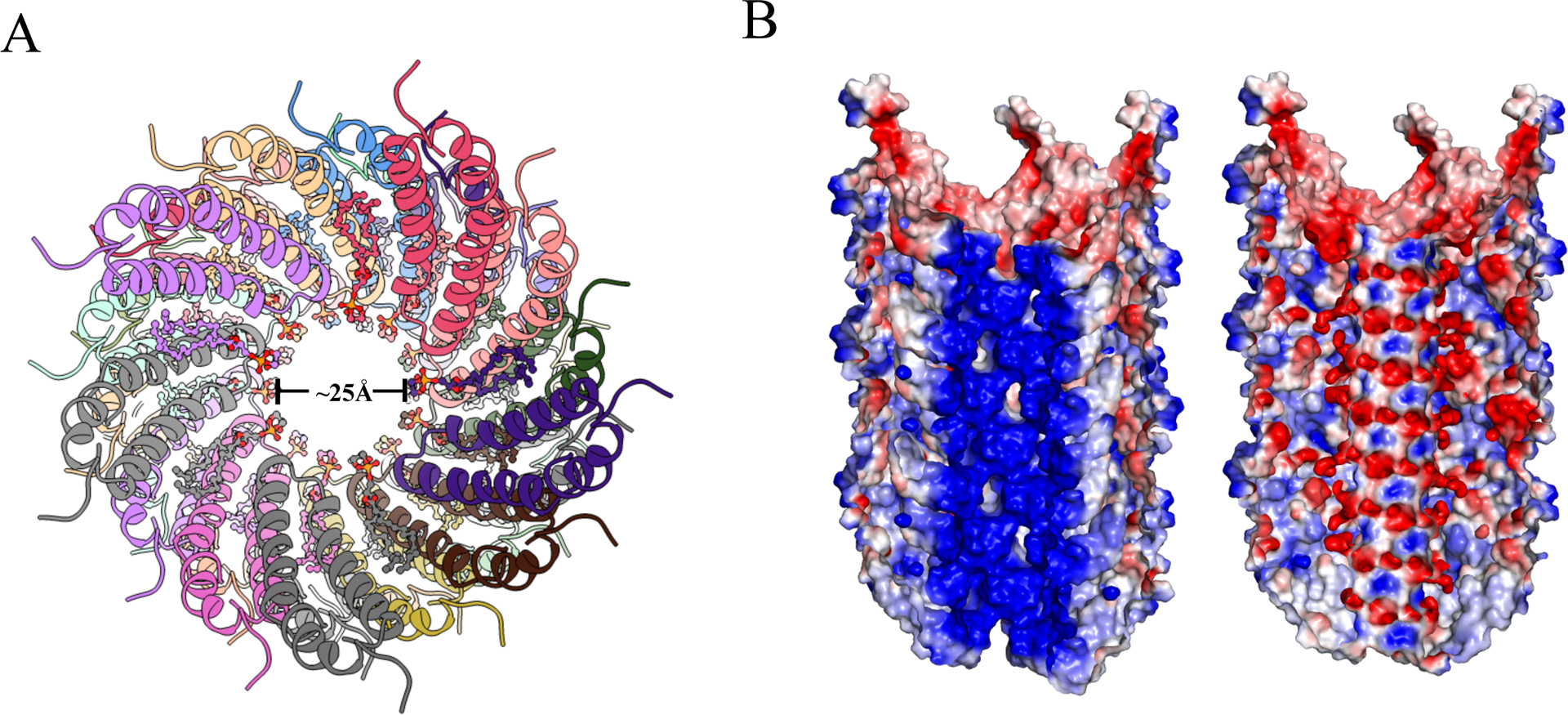
(A) Top view of cartoon representation of pKpQIL pili, showing the head group of the phospholipid pointing into the lumen. The phospholipid is shown in a ball & stick representation. (B) Cut-away view of electrostatic surface of phospholipid-free pKpQIL pili (left) and phospholipid-bound pKpQIL pili (right) show how the phosphate groups lining the lumen make the surface electronegative. Surface representation colored by electrostatic potential ranging from −3 kT/e (red) to 3 kT/e (blue).
Discussion
Infections with carbapenem-resistant K. pneumoniae pose a significant public health concern, particularly in healthcare setting, where the requirement for new therapeutics to treat antibiotic-resistant bacterial infections is critical (Tacconelli et al., 2017). Since conjugative pili play an important role in the horizontal transfer of antibiotic-resistance genes, understanding the structure of these pili might generate druggable new targets. For example, the molecular structure of conjugative pili can be employed in engineering specific bacteriophages to treat bacterial infection through targeting these pili (Chen et al., 2019).
We describe the cryo-EM analysis of the conjugative pili encoded by the pKpQIL plasmid, and our results reveal a different helical packing (Fig. S3) from the F-type pED208 pilus or the F pOX38 pilus that have 5-fold rotational symmetry (Costa et al., 2016). Despite this difference in helical symmetry, the pilin folds are similar between the two forms of conjugative pili. However, the intermolecular interfaces are different. A major question is the biological relevance of these two distinct packing arrangements. One possibility is that the differences in packing are partly or largely responsible for phage recognition. If this were the case, then phages might be supplying selective pressure driving the structural diversification. Filamentous ssDNA phages (M13, fd, f1) can bind to the tip of F-type pili and the icosahedral single-stranded RNA MS2-like phages, including MS2 and R17, can bind to the side of the F-type pili (Manchak et al., 2002). The distinct helical packing of F-type pili might explain the variability in their binding affinity to a variety of phages. The F-pilus was reported to form two electrostatic interactions with the MS2 Mat protein through its N-terminus (Meng et al., 2019). Mutations in the N-terminus have been shown to significantly reduce the affinity of specific phages to the F-pilus.
An interesting question is whether the structural class of the mating pili can be determined from sequences. Sequence analysis reveals that pKpQIL is closer to F (pOX38) than to pED208 (Figure 3A,B), but both the pED208 and pOX38 have the same 5-fold symmetry (Costa et al., 2016). Thus, with only one example of a mating pilus with a 1-start helix (pKpQIL) and two examples of mating pili with 5-fold symmetry (pED208 and pOX38) simple sequence comparisons fail to show the proper classification and many more structures will be needed.
It has previously been shown that relatively small perturbations can change the quaternary structure of helical polymers without any significant changes to the secondary or tertiary structure of the assembling subunits. For example, a 29-residue peptide polymerizes into a very different helical polymer after a semi-conservative change of a single lysine to an arginine (Egelman et al., 2015). Pyrin domains that form an important part of the inflammasome were shown to polymerize into filaments with 3-fold symmetry (Lu et al., 2014). However, when a pyrin domain was tagged with GFP, the filaments had a 1-start symmetry due to steric conflicts between the GFP tags, showing the plasticity in the helical packing (Lu et al., 2015). We have shown here that the different helical symmetry found for pKpQIL compared to the previous structures of pED208 and pOX38 represents an amplification of differences in the primary sequences by different quaternary interactions.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Edward Egelman (egelman@virginia.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The atomic has been deposited at the Protein Data Bank (www.rcsb.org) with accession code 7JSV. The corresponding cryo-EM map was deposited in the Electron Microscopy Data Bank (www.ebi.ac.uk/pdbe/emdb) with accession code EMD-22460.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The samples were prepared from K. pneumoniae pKpQILΔfinO.
METHOD DETAILS
Sample preparation
Two litre cultures of K. pneumoniae pKpQILΔfinO were grown overnight in LB at 37°C. Cells were harvested by centrifugation at 7,000 × g for 20 min. Pellets were resuspended in 40 ml of cold PBS and passed 30 times through a 25G needle. “Shaved” bacteria were centrifuged at 50,000 × g for 1 h, and the supernatant was collected, mixed with 5% PEG6000, incubated for 1 h at 4°C and centrifuged at 50,000 × g for 30 min. Precipitated pili were resuspended in a buffer containing 50 mM Tris pH8, 1 M NaCl and dialysed over-night against the same buffer. Dialysed material was concentrated in a 30 kDa Centricon.
Cryo-EM data collection and processing
2 μl of pKpQIL pili were applied to plasma-cleaned lacey carbon grids, followed by plunge-freezing in liquid ethane using a Leica EM GP. Data collection was carried out at liquid nitrogen temperature on a Titan Krios microscope (Thermo Fisher Scientific) operated at an accelerating voltage of 300 kV. 40 movie frames were collected with defocus values range between −1 to −2.5 μm on a K3 camera (Gatan), with a total dose of ~55 electrons per Å2. A total of 263,265 256 px-long segments were manually boxed in EMAN2 (Tang et al., 2007) from 516 images, followed by importing and extracting in RELION 3.0 software (Zivanov et al., 2018). Out of 50 two-dimensional classes, 9 classes (containing 163,128) segments were selected for Refine3D, which yield a map of recognizable secondary structure features when imposing 1-start helical symmetry. The helical parameters converged to a rotation of 77.6° and an axial rise of 2.7 Å per subunit. After contrast transfer function (CTF) refinement and Bayesian polishing, the resolution of the final reconstruction was determined by the Fourier shell correlation (FSC) between two independent half maps, which was 3.9 Å at FSC = 0.143 (Figure S2).
Model building and refinement
We used the pED208 pili (PDB ID: 5LEG) as an initial template to dock into the cryo-EM map by rigid body fitting, and then manually edited the model in UCSF Chimera (Pettersen et al., 2004) and Coot (Emsley et al., 2010). The modified model was used as the starting template to further refine with RosettaCM (Song et al., 2013). The refined monomeric model of pKpQIL pili was then re-built by RosettaCM with helical symmetry, followed by real-space refinement with Phenix (Adams et al., 2010). The final model was validated by MolProbity (Chen et al., 2010). The data collection and refinement statistics are listed in Table 1. To calculate the electrostatic potential of the phospholipid-free pili as well as the phospholipid-bound pili, corresponding PDB format files were converted to PQR format with the PDB2PQR server (Dolinsky et al., 2007) using the PEOEPB force field and then applied to the APBS program (Baker et al., 2001). Structure figures were created using UCSF Chimera and Pymol.
Table 1.
Cryo-EM Data collection, processing and model refinement statistics of the pKpQIL pilus
| Data collection and processing | |
| Microscope | Titan Krios |
| Camera | K3 |
| Voltage (kV) | 300 |
| Electron exposure (e− Å −2) | 55 |
| Pixel size (Å) | 1.08 |
| Defocus range (uM) | −1.0~ −2.5 |
| Helical rise (Å) | 2.7 |
| Helical twist (°) | 77.6 |
| Map resolution (Å) | 3.9 |
| Map B-factor | −170 |
| Refinement and Model validation | |
| Bond lengths rmsd (Å) | 0.008 |
| Bond angles rmsd (°) | 0.911 |
| Clashscore | 6.73 |
| Poor rotamers (%) | 0 |
| Ramachandran Favored (%) | 100 |
| Ramachandran Outlier (%) | 0 |
| MolProbity score | 1.37 |
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was only relevant for the atomic model, and those results are presented in Table 1.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| pKpQILΔfinO | Frankel lab, Imperial College | pICC4002 |
| pKpQIL pili | Frankel lab, Imperial College | pKpQIL pili |
| Deposited Data | ||
| pKpQIL conjugative pili map | This paper | EMD-22460 |
| pKpQIL conjugative pili model | This paper | PDB: 7JSV |
| pED208 conjugative pili model | Costa et al., 2016 | PDB:5LEG |
| Software and Algorithms | ||
| MotionCor2 | Zheng et al., 2017 | https://emcore.ucsf.edu/ucsf-motioncor2 |
| EMAN2 | Tang et al., 2007 | https://blake.bcm.edu/emanwiki/EMAN2 |
| RELION3 | Zivanov et al., 2018 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page |
| CHIMERA | Goddard et al., 2007 | https://www.cgl.ucsf.edu/chimera/ |
| COOT | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| ROSETTA | Song et al., 2013 | https://www.rosettacommons.org/software |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org/ |
Highlights.
A 3.9 Å resolution cryo-EM structure for a bacterial mating pilus
Unlike two related mating pili, this filament does not have 5-fold symmetry
Filamentous phage have also been described as existing in two structural classes
Small changes in sequence can have large changes on quaternary structure
Acknowledgements
Cryo-EM images were collected at the University of Virginia Molecular Electron Microscopy Core facility, which is supported by the School of Medicine and built with NIH grant G20-RR31199. Work in the EHE lab is supported by NIH grant GM122510. Work in the GF lab is supported by a Wellcome Trust Investigator award 107057/Z/15/Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony KG, Klimke WA, Manchak J, and Frost LS (1999). Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100–1: insights into the mechanism of conjugation. J Bacteriol 181, 5149–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony KG, Sherburne C, Sherburne R, and Frost LS (1994). The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol 13, 939–953. [DOI] [PubMed] [Google Scholar]
- Arutyunov D, and Frost LS (2013). F conjugation: back to the beginning. Plasmid 70, 18–32. [DOI] [PubMed] [Google Scholar]
- Babic A, Lindner AB, Vulic M, Stewart EJ, and Radman M (2008). Direct visualization of horizontal gene transfer. Science 319, 1533–1536. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, and McCammon JA (2001). Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran Darbari V, and Waksman G (2015). Structural biology of bacterial type IV secretion systems. Annual review of biochemistry 84, 603–629. [DOI] [PubMed] [Google Scholar]
- Chen L, Chavda KD, Melano RG, Jacobs MR, Koll B, Hong T, Rojtman AD, Levi MH, Bonomo RA, and Kreiswirth BN (2014a). Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob Agents Chemother 58, 2871–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, and Kreiswirth BN (2014b). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends in microbiology 22, 686–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica Section D, Biological crystallography 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Batra H, Dong J, Chen C, Rao VB, and Tao P (2019). Genetic Engineering of Bacteriophages Against Infectious Diseases. Front Microbiol 10, 954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Maddera L, Harris RL, and Silverman PM (2008). F-pili dynamics by live cell imaging. Proceedings of the National Academy of Sciences of the United States of America in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TR, Ilangovan A, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, and Waksman G (2016). Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex. Cell 166, 1436–1444 e1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daehnel K, Harris R, Maddera L, and Silverman P (2005). Fluorescence assays for F-pili and their application. Microbiology 151, 3541–3548. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, and Baker NA (2007). PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic acids research 35, W522–W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH (2014). Ambiguities in Helical Reconstruction. eLife 3:e04969 doi: 10.7554/eLife.04969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH, Xu C, DiMaio F, Magnotti E, Modlin C, Yu X, Wright E, Baker D, and Conticello VP (2015). Structural plasticity of helical nanotubes based on coiled-coil assemblies. Structure 23, 280–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta crystallographica Section D, Biological crystallography 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Ippen-Ihler K, and Skurray RA (1994). Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev 58, 162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman TH, Frost LS, and Silverman PM (1990). Structure and function of conjugative pili: monoclonal antibodies as probes for structural variants of F pili. J Bacteriol 172, 1174–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RL, and Silverman PM (2004). Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J Bacteriol 186, 5480–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, and Rosenstrom P (2010). Dali server: conservation mapping in 3D. Nucleic Acids Res 38, W545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, and Lee SH (2016). Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front Microbiol 7, 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WW, Wong J, Pena A, Seddon C, Costa T, Beis K, and Frankel G (2020). OmpK36 and TraN facilitate conjugal transfer of the Klebsiella pneumoniae carbapenem resistance plasmid pKpQIL. bioRxiv, 2020.2007.2001.180638. [Google Scholar]
- Lu A, Li Y, Yin Q, Ruan J, Yu X, Egelman EH, and Wu H (2015). Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discovery 1, 15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, and Egelman EH (2014). Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell 156, 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmborg AC, Soderlind E, Frost L, and Borrebaeck CA (1997). Selective phage infection mediated by epitope expression on F pilus. J Mol Biol 273, 544–551. [DOI] [PubMed] [Google Scholar]
- Manchak J, Anthony KG, and Frost LS (2002). Mutational analysis of F-pilin reveals domains for pilus assembly, phage infection and DNA transfer. Mol Microbiol 43, 195–205. [DOI] [PubMed] [Google Scholar]
- Meng R, Jiang M, Cui Z, Chang JY, Yang K, Jakana J, Yu X, Wang Z, Hu B, and Zhang J (2019). Structural basis for the adsorption of a single-stranded RNA bacteriophage. Nature communications 10, 3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva WD, and Silverman PM (1996). Effects of F-encoded components and F-pilin domains on the synthesis and membrane insertion of TraA’-’PhoA fusion proteins. Mol Microbiol 19, 1277–1286. [DOI] [PubMed] [Google Scholar]
- Panicker MM, and Minkley EG Jr. (1985). DNA transfer occurs during a cell surface contact stage of F sex factor-mediated bacterial conjugation. Journal of Bacteriology 162, 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. JComputChem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Silverman PM (1997). Towards a structural biology of bacterial conjugation. MolMicrobiol 23, 423–429. [DOI] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, and Baker D (2013). High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E, Magrini N, Kahlmeter G, and Singh N (2017). Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization 27, 318–327. [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. Journal of Structural Biology 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Waksman G (2019). From conjugation to T4S systems in Gram-negative bacteria: a mechanistic biology perspective. EMBO reports 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Dayan N, Goldbourt A, and Xiang Y (2019). Cryo-electron microscopy structure of the filamentous bacteriophage IKe. Proc Natl Acad Sci U S A 116, 5493–5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Wang F, Taylor NMI, Guerrero-Ferreira RC, Leiman PG, and Egelman EH (2017). Refined Cryo-EM Structure of the T4 Tail Tube: Exploring the Lowest Dose Limit. Structure 25, 1436–1441 e1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic has been deposited at the Protein Data Bank (www.rcsb.org) with accession code 7JSV. The corresponding cryo-EM map was deposited in the Electron Microscopy Data Bank (www.ebi.ac.uk/pdbe/emdb) with accession code EMD-22460.


