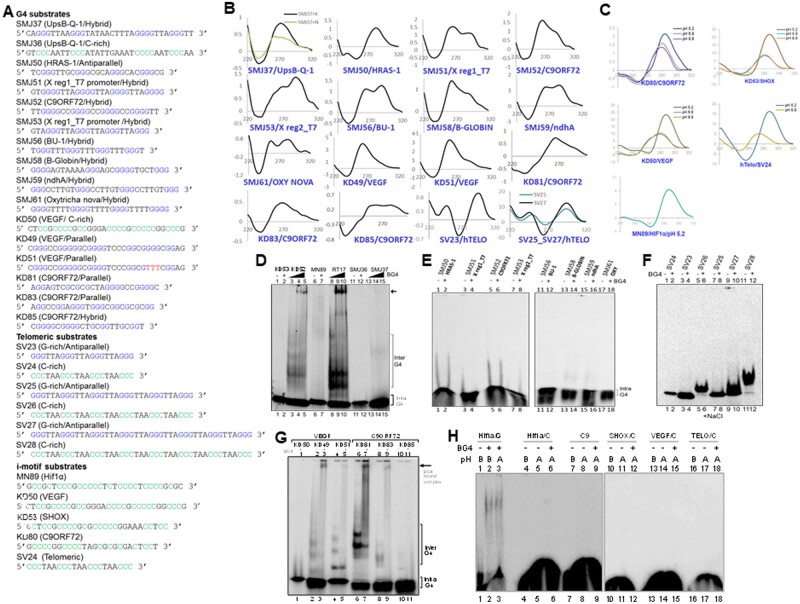
Figure 4.
Evaluation of BG4 binding to different orientations of G-quadruplex DNA structures. (A) Sequences of oligomeric DNA substrates used for the study. (B) CD spectra showing G4 formation in parallel, antiparallel and hybrid orientation. DNA sequences used for studying G4 in parallel orientation were, SMJ50 (HRAS-1), KD49 (VEGF), KD51 (VEGF), KD81 (C9ORF72) and KD83 (C9ORF72). G4 structures studied in antiparallel orientation (in presence of 100 mM NaCl) were, oligomeric DNA-containing telomeric sequences SV23, SV25 and SV27. Oligomers SMJ37 (UpsB-Q-1), SMJ51 (X Reg1/T7 promoter), SMJ52 (C9ORF72), SMJ53 (X Reg2/T7 promoter), SMJ56 (BU-1), SMJ58 (B-GLOBIN), SMJ59 (ndhA), SMJ61 (OXY) showed more of hybrid G4 structures with dominant antiparallel orientation (in presence of 100 mM NaCl). For other details, refer Fig. 3B. (C) CD spectra showing i-motif formation in presence of 100 mM potassium acetate (pH 5.2), Tris buffer (pH 6.8) and Tris buffer (pH 8.8), at room temperature. Reading was taken from wavelength range of 220–320 nm (10 cycles), using a spectropolarimeter (JASCO J-810) at scan speed of 50 nm/min. Absorption spectrum measured for buffer alone (15 cycles) was subtracted from each sample and resulting spectra were plotted. In case of all the sequences (KD80, KD50, SV24, KD53, MN89), acidic pH of 5.2 promoted i-motif formation, as seen from positive peak at ∼285 nm and dip at ∼255 nm. (D) Comparison of BG4-binding efficiency between oligomeric DNA that can form parallel and antiparallel G-quadruplexes. Parallel G-quadruplex forming oligomers RT17 (Hif1α; lanes 8-10), KD52 (SHOX; lanes 3-5) and antiparallel G-quadruplex-forming oligomers SMJ37 (UpsB-Q-1; lanes 13-15) were incubated with increasing concentrations of BG4 (0.0, 0.4, 0.8 µg) and resolved on PAGE. In every case complementary DNA (KD53, lanes 1, 2; MN89, lanes 6, 7; SMJ36, lanes 11, 12) was used as control for BG4 binding. (E) Comparison of BG4-binding efficiency between oligomeric DNA that can form antiparallel G-quadruplexes. Antiparallel G-quadruplex forming oligomers SMJ50 (HRAS-1; lanes 1–2), SMJ51 (X Reg1/T7 promoter, lanes 3–4), SMJ52 (C9ORF72, lanes 5–6), SMJ53 (X Reg2/T7 promoter, lanes 7–8), SMJ56 (BU-1, lanes 11–12), SMJ58 (B-GLOBIN, lanes 13–14), SMJ59 (ndhA, lanes 15–16), SMJ61 (OXY, lanes 17–18) were incubated with BG4 (1.0 µg) and resolved on 6% native PAGE. (F) Assessment of BG4 binding to oligomers containing antiparallel G4 structures (SV23, lanes 3–4; SV25, lanes 7–8; SV27, lanes 9–10) and their complementary sequences (SV24, lanes 1–2; SV26, lanes 5–6; SV28, lanes 11–12). Substrates were incubated at 37°C in presence of 100 mM sodium chloride, followed by incubation with 1.5 µg of BG4 at 4°C, and the resulting complexes were resolved on a 6% native PAGE containing 100 mM NaCl. (G) Evaluation of binding efficiency of G4 DNA derived from VEGF and C9ORF72 regions, containing GNG motifs. Oligomers were incubated at 37°C in presence of 100 mM KCl, followed by incubation with 1.0 µg of BG4 at 4°C, and products were resolved on a 6% native PAGE. (H) Comparison of BG4-binding efficiency between DNA substrates containing G4 structure, RT17 (HIF1Α; lanes 1-3) and DNA containing i-motifs, MN89 (HIF1α; lanes 4–6), KD80 (C9ORF72; lanes 7–9), KD52 (SHOX; lanes 10-12), KD50 (VEGF; lanes 13–15) and SV24 (telomeric region; lanes 16–18). RT17 was incubated at 37°C, in presence of 100 mM KCl, whereas i-motif substrates were incubated with 100 mM potassium acetate (pH 5.2) at 95°C for 2 h and cooled to room temperature.