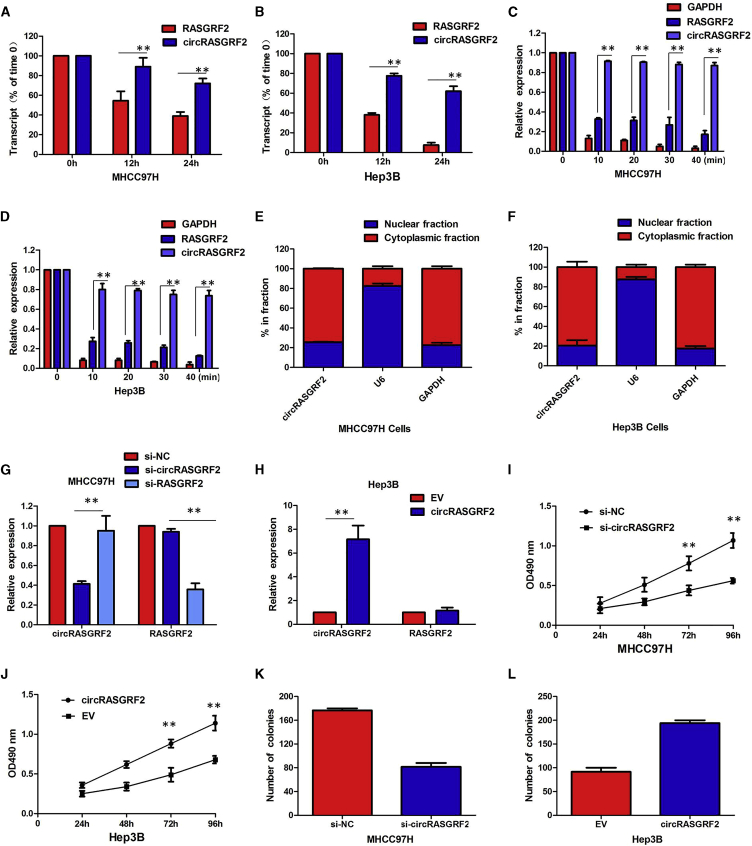
Figure 2.
circRASGRF2 loss of function dramatically impairs the HCC cell malignant phenotypes in vitro
(A) Quantitative real-time PCR for the abundance of circRASGRF2 and RASGRF2 in MHCC97H cells treated with actinomycin D at the indicated time point. (B) Quantitative real-time PCR for the abundance of circRASGRF2 and RASGRF2 in Hep3B cells treated with actinomycin D at the indicated time point. (C) Quantitative real-time PCR for the expression of circRASGRF2 and RASGRF2 mRNA in MHCC97H cells treated with or without RNase R. (D) Quantitative real-time PCR for the expression of circRASGRF2 and RASGRF2 mRNA in Hep3B cells treated with or without RNase R. (E) Levels of circRASGRF2 in the nuclear and cytoplasmic fractions of MHCC97H cells. (F) Levels of circRASGRF2 in the nuclear and cytoplasmic fractions of Hep3B cells. (G) The expression of circRASGRF2 was only downregulated by si-circRASGRF2 but was not affected by si-RASGRF2. (H) The circRASGRF2 expression vector markedly increased the expression of circRASGRF2 compared with the empty vector. (I) CCK-8 assays showed circRASGRF2 knockdown significantly decreased the cell viability of MHCC97H cells. (J) Overexpression of circRASGRF2 promoted the proliferative ability of Hep3B cells. (K) Colony formation assays revealed that circRASGRF2 knockdown greatly attenuated the numbers of visible colonies of MHCC97H cells. (L) Colony formation assays revealed that overexpression of circRASGRF2 greatly increased the numbers of visible colonies of Hep3B cells. All tests were performed at least three times. Data are expressed as mean ± SD. ∗∗p < 0.01, ∗∗∗p < 0.001.