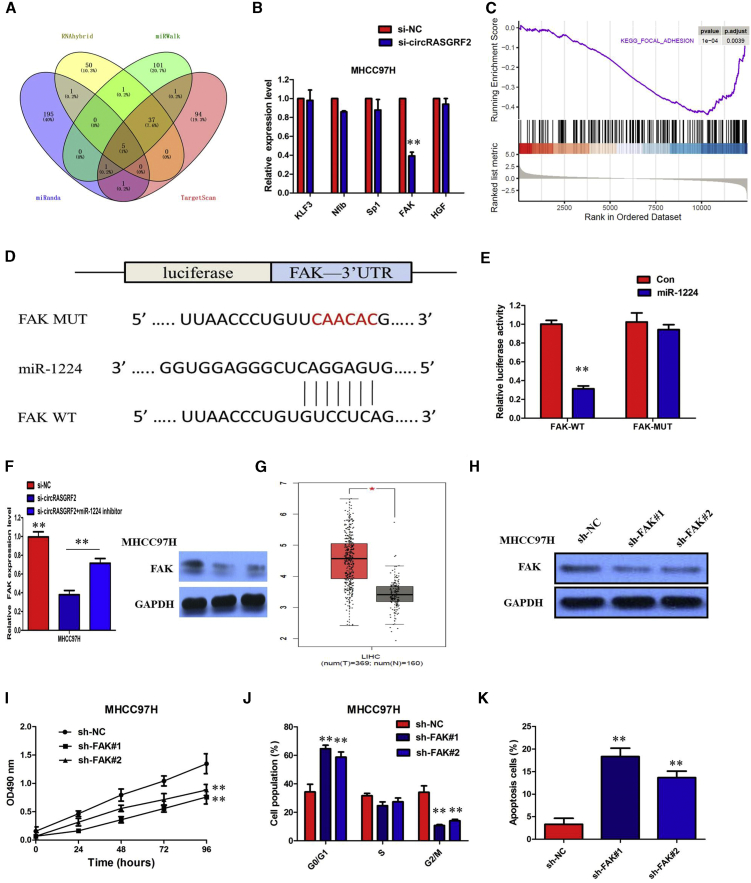
Figure 7.
FAK was positively regulated by circRASGRF2/miR-1224
(A) Venn diagram showing five genes that are putative miR-1224 targets computationally predicted by four algorithms (miRanda, RNAhybrid, miRWalk, and TargetScan). (B) mRNA levels of five candidate target genes were detected after silencing circRASGRF2. (C) GSEA analysis showed that the gene set of KEGG_FOCAL ADHESION_PATHWAY was significantly enriched in the si-circRASGRF2 group compared to the si-NC group. (D) Binding sequence between miR-1224 and FAK. (E) Luciferase reporter assay demonstrated that miR-1224 mimics significantly decreased the luciferase activity of FAK-WT in HEK293 T cells. (F) Inhibition of the circRASGRF2-mediated decrease of FAK mRNA and protein expression was significantly recuperated following miR-1224 inhibitors. (G) TCGA database showed that the level of FAK was significantly upregulated in HCC tissue compared with normal liver tissue. (H) The expression of FAK was downregulated in MHCC97H cells by two shRNAs. (I) MHCC97H cells transduced with FAK shRNAs had a slower growth rate. (J) The cell cycle analysis showed that more MHCC97H cells were distributed in the G1 phase and less in the S phase after silencing FAK. (K) FAK knockdown significantly increased the percentage of apoptotic cells in MHCC97H cells. All tests were performed at least performed three times. Data are expressed as mean ± SD. ∗∗p < 0.01.