Figure 3.
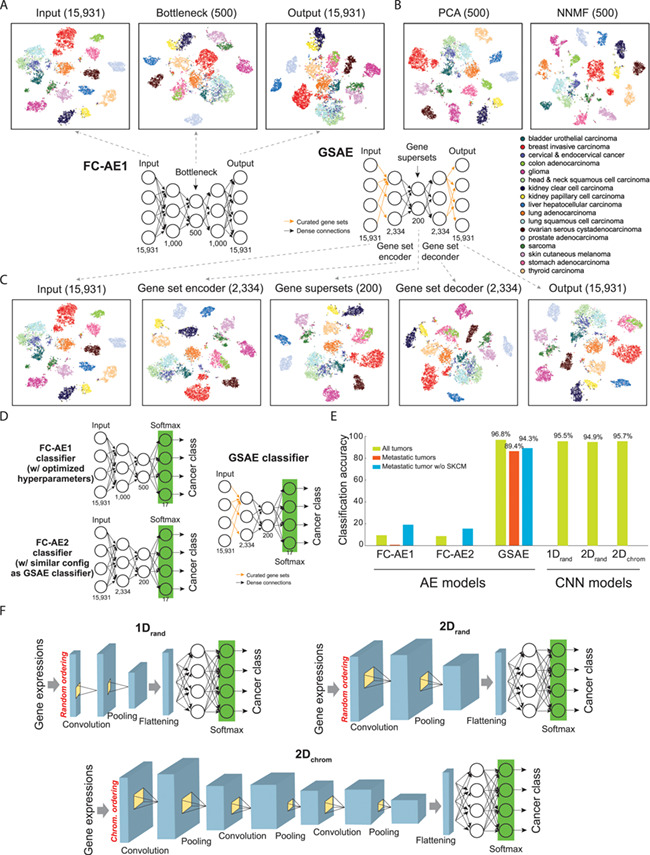
Classifying cancer types using pan-cancer gene expression data. (A) Dimension reduction by fully connected AEs (FC-AE) of 17 cancer types. (B) Dimension reduction by PCA and NNMF. (C) Architecture of GSAE that incorporates curated gene sets into an AE and t-SNE visualization of outputs at each layer. (D) Architectures of models classifying cancer types of primary and metastatic tumors by linking the bottleneck layer of AEs to a classification layer. Performance of a GSAE classifier was compared to two AEs, an FC-AE1 classifier with hyperparameter optimization (as shown in A) and an FC-AE2 classifier that introduced full connections to the GSAE classifier. (E) Performance of AE and CNN-based classifiers of cancer types. Performance of FC-AE1, FC-AE2 and GSAE was assessed by our in-house analysis of 17 largest cancer types using 10-fold cross validation. Performance of 1Drand and 2Drand was reported by [58] on all 33 cancer types by 5-fold cross validation; 2Dchrom was evaluated by [59] on all 33 cancer types by a 10-fold cross validation, where subscript ‘chrom’ denotes genes ordered by chromosomal position and ‘rand’ for genes randomly ordered. (F) Architectures of different embedding methods of gene expression data and CNN models for classifying cancers proposed by [58] and [59].
