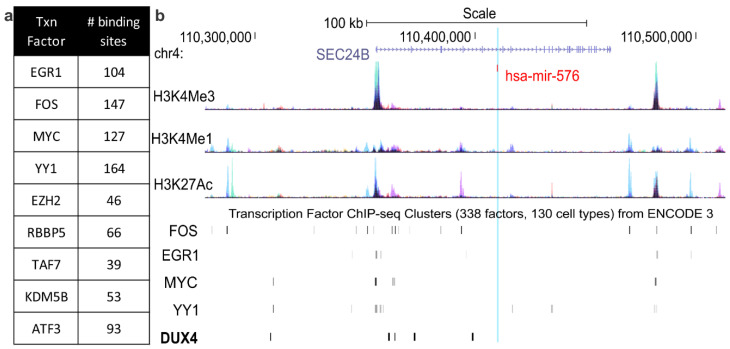
Figure 2.
Candidate miRNA loci are consistent with regulation via transcription factors dysregulated in FSHD. (a) Table listing a subset of transcription factors which are each increased in human skeletal muscle cells in response to DUX4 overexpression [9], along with the number (#) of binding sites they show within potential regulatory distance (100 kb) of the 19 candidate miRNAs. (b) The miR-576 locus shows binding consistent with regulation by FOS, EGR1, MYC, YY1, and DUX4. Corresponding epigenetic modification maps display the location of histone modifications associated with active promoters (H3K4me3) and poised/active enhancers (H3K4me1 and H3K27Ac) in the vicinity of the miR-576 locus and its surrounding home gene, SEC24 homolog B (SEC24B). (DUX4 binding sites identified using ChIP-seq data uploaded from Geng et al. [9]; binding sites for additional transcription factors identified using UCSC Genome Browser and respective ChIP-seq datasets accessed via the ENCODE3 regulation track [46,47,48,49,50]).