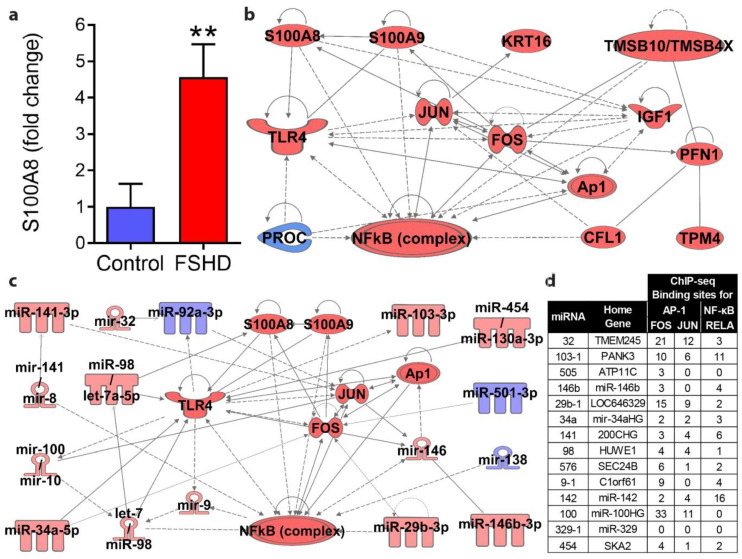
Figure 5.
Validation and pathway analysis of elevated S100A8 protein in FSHD. (a) ELISA of S100A8 protein in plasma from a separate validation set of FSHD1 patients. (b) Bioinformatic pathway analysis was used to identify known connections between candidate protein markers with S100A8 pathway proteins involved in TLR4 signaling. (c) Bioinformatic pathway analysis was used to identify established connections between candidate miRNAs with S100A8 pathway proteins involved in TLR4 signaling. (d) Bioinformatic analysis of ChIP-seq defined binding sites for the key S100A8 pathway transcription factors AP-1 (FOS and JUN) and NF-κB (RELA), at potential regulatory regions of the candidate miRNAs that were found to increase in FSHD plasma. Binding sites represent the combined number of potential promoter (within 2 kb of promoter) and enhancer (within 10 kb) regulatory regions with ChIP-seq-confirmed transcription factor binding for each miRNA home gene. (** p ≤ 0.01; n = 13 healthy control volunteers, 19 FSHD1; panels (b,c) produced using Ingenuity Pathway Analysis software, red = increased, blue = decreased; data for panel (d) produced using the Factorbook ChIP-seq data repository from ENCODE and the UCSC genome browser).