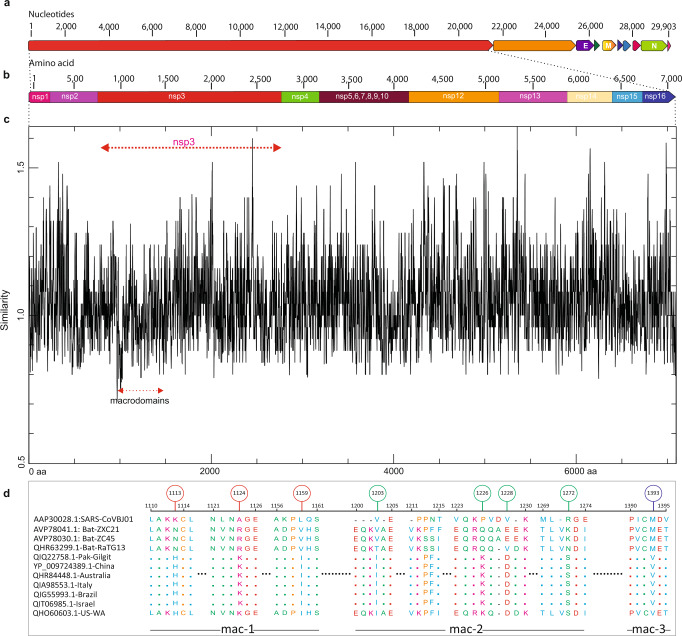
Fig. 2. Identification of sequence divergence at the polyprotein pp1ab locus in SARS-CoV-2.
a Schematic of SARS-CoV-2 genome. Genomic organization of SARS-CoV-2 with numbering above the block referring to nucleotide positions. Structural proteins, including spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, as well as nonstructural proteins (nsps) translated from ORF1ab and accessory proteins are indicated. b Schematic of 7096-aa replicase polyprotein pp1ab with its 15 sub-proteins (nsp1–nsp10 and nsp12–nsp16). c Comparative sequence analysis of the pp1ab domain for completely sequenced genomes of SARS-CoV-2 from at least 39 distinct global territories. SARS-CoV-2 sequences were compared to the corresponding homologous sequence from batSL-CoVs and SARS-CoVBJ01. Macrodomains within Nsp3 are demarcated by red arrow. Y axis depicts similarity scores between CoVs and the X axis refers to the relative residue position. Lower scores signify low sequence conservation, with trough corresponding to the least conserved regions. d Macrodomains sequences encoding Mac-1, Mac-2, and Mac-3 proteins exhibiting exceptional divergence in SARS-CoV-2 relative to bat-RaTG13/ZC45/ZXC21 and SARS-CoVBJ01. Green, red, and blue circles, respectively, differentiate between Mac-1, Mac-2, and Mac-2 specific substitutions.